|
Inheritance of resistance to the root-knot nematode Meloidogyne javanica in lettuce
ABSTRACT. Resistance to the root-knot nematodes Meloidogyne spp. would be a valuable attribute of lettuce Lactuca sativa L. cultivars grown in tropical regions. The looseleaf lettuce ‘Grand Rapids’ is resistant to both M. incognita and M. javanica. Resistance to M. incognita has a high heritability, under the control of a single gene locus, in which the ‘Grand Rapids’ allele, responsible for resistance (Me), has predominantly additive gene action, and has incomplete penetrance and variable expressivity. We studied the inheritance of the resistance of ‘Grand Rapids’ (P2) to M. javanica in a cross with a standard nematode-susceptible cultivar Regina-71 (P1). F1 (Regina-71 x Grand Rapids) and F2 seed were obtained, and the F2 inoculated, along with the parental cultivars, with a known isolate of M. javanica to evaluate nematode resistance. A high broad sense heritability estimate (0.798) was obtained for gall indices. Class distributions of gall indices for generations P1, P2, and F2 were in agreement with theoretical distributions based on a monogenic inheritance model for the range of assumed degrees of dominance between approximately -0.20 and 0.20. M. javanica resistance appears to be under control of a single gene locus, with predominantly additive gene action. Whether or not the Grand Rapids allele imparting resistance to M. javanica is the same Me allele imparting resistance to M. incognita remains to be determined. Key words: Lactuca sativa L., Lettuce, Resistance, Meloidogyne javanica, Nematode
INTRODUCTION Varietal resistance is considered one of the most efficient methods for root-knot nematode (Meloidogyne spp.) control (Ferraz and Mendes, 1992; Maluf, 1997); however, little emphasis has been placed on breeding lettuce for nematode resistance. In Brazil, both M. javanica and M. incognita are widespread (Lordello, 1984) and are known to occur sympatrically. Multiple resistance to both species and their races (in the case of M. incognita) is an important breeding goal. In lettuce, root-knot nematodes were not recognized as a serious problem until recently (Campos, 1995). Reports of nematode attacks have become more common with increasing lettuce acreages grown under plastic tunnels, leading to increases in nematode populations. The use of susceptible crisphead cultivars, whose cycle is usually much longer than that of traditionally grown butterhead or looseleaf cultivars, has also impacted on nematode populations. Lettuce breeding practices in Brazil have emphasized selection of genotypes with good vegetative growth under high temperatures, and resistance to the lettuce mosaic virus (LMV) (Nagai and Costa, 1972, 1973; Costa and Silva, 1976; Nagai, 1979, 1980, 1983; Nagai and Lisbão, 1980; Silva et al., 1999). Resistance to Meloidogyne spp. in lettuce received little or no attention in the past, but is now considered an important goal in breeding lettuce for both field and greenhouse production in Brazil (Mendes, 1998; Gomes, 1999). There is little published on varietal resistance to Meloidogyne spp. in lettuce. Ryder’s comprehensive review of lettuce genetics and breeding (Ryder, 1986) does not report the existence of root-knot nematode resistant lettuce germplasm. Kaloo (1988) reviewed sources of Meloidogyne resistance in vegetable crop species, but made no reference to lettuce. An evaluation of the USDA and the University of California Lactuca spp. germplasm collections (McGuire et al., 1993) similarly made no reference to sources of resistance to root-knot nematodes. Charchar (1991) was apparently the first to report Meloidogyne resistance in lettuce. This report was later confirmed by Gomes et al. (1996), Charchar and Moita (1996), and Mendes (1998). Resistance to both M. incognita (races 1, 2, 3 and 4) and M. javanica was found in the Grand Rapids cultivar (Mendes, 1998; Gomes, 1999). Gomes (1999) found that the resistance of the lettuce cultivar Grand Rapids to M. incognita has high heritability, and is under control of a single gene locus in which the resistance allele shows incomplete penetrance, variable expression and predominantly additive action. A symbol (Me) was proposed for the cultivar Grand Rapids allele that imparts resistance to M. incognita, but inheritance of this resistance to M. javanica has not been studied. We studied the genetic control of resistance to the root-knot nematode species M. javanica in the lettuce cultivar Grand Rapids. MATERIAL AND METHODS Plant material Two lettuce cultivars - Grand Rapids and Regina-71, with contrasting reactions to root-knot nematodes, were used in this study. Regina-71 is a Brazilian white-seeded butterhead cultivar previously characterized as susceptible to both M. incognita and M. javanica (Gomes et al., 1996; Mendes, 1998; Gomes, 1999). Grand Rapids is a black-seeded looseleaf American cultivar, previously found to be resistant to both root-knot nematode species (Gomes et al.,1996; Mendes, 1998; Gomes, 1999). Crosses between Regina-71 (seed parent) and Grand Rapids (pollen parent) were made in accordance with standard emasculation and pollination techniques (Nagai, 1980; Ryder, 1986). Seed color was used as a marker to unequivocally identify true F1 plants (Ryder, 1986): putative F1 plants were grown to seed production, and any white-seeded seed parent contaminant plants were eliminated, leaving only true F1 plants from which F2 seed were secured. Plants of parental generations P1 (= Regina-71), P2 (= Grand Rapids), F2 (Regina-71 x Grand Rapids) were used to study the inheritance of nematode resistance. The F1 (Regina-71 x Grand Rapids) generation was not included in the trial, due to an insufficient supply of F1 seed. Inoculum preparation and inoculation methods A known isolate of M. javanica (characterized at the Department of Plant Pathology, Universidade Federal de Lavras, Lavras, MG, Brazil) was routinely maintained on isolated benches within plastic houses at the Vegetable Research Station of HortiAgro Sementes Ltda., Ijaci, MG, Brazil. Tomato plants of the susceptible cultivar Santa Clara were grown in 8-liter pots and inoculated with 10,000 nematode eggs/pot. Nematode inocula were prepared in accordance with the technique described by Hussey and Barker (1973), modified by Boneti (1981). Approximately 70 days after inoculation in tomato plants, nematode eggs were extracted by the same technique, and used as inocula for the lettuce plants. A 50% v/v mixture of commercial substrate (PlantimaxÒ) + carbonized rice husks was inoculated with the nematode egg suspension, homogenized to a final concentration of 30 eggs per ml mixture, and used to fill styrofoam speedling trays with trapezoidal cells (35 ml substrate per cell). Lettuce seedlings at the one-true-leaf stage were transplanted into the trays (one seedling/cell), where they were kept until the date of resistance evaluation. Evaluation of plant reactions to M. javanica The evaluation date was chosen based on the presence of abundant numbers of galls in the root system of plants of the susceptible cultivar Regina-71. Inoculated lettuce plants grown in speedling cells had their root systems washed in tap water before scoring for gall indices (GI). GI were assessed on a scale of 1 to 5, based on the number of galls per root system and average gall diameter, as follows: 1 = no galls, or sparse galls with average gall diameter £1 mm; 2 = sparse galls, with average gall diameter between 1 and 2 mm; 3 = galls mostly non-coalescent, average gall diameter between 2 and 3 mm; 4 = galls numerous and coalescent, average gall diameter between 3 and 4 mm, and 5 = galls numerous and coalescent, average gall diameter >4 mm. Resistance trials with parental and F2 generations A resistance trial was carried out with the parental cultivars Regina-71 (= P1) and Grand Rapids (= P2), and the segregating F2 (Regina-71 x Grand Rapids) generation from November 1998 through February 1999. Plants were distributed in 8-plant plots, whose positions were assigned to the trays in accordance with a completely randomized design. The total number of plants evaluated (final stand) was 99, 118 and 484, for P1, P2 and F2, respectively. Inoculation procedures were as previously described. Plant reactions to the nematode were evaluated 70 days after transplanting/inoculation. During the period prior to evaluation of nematode resistance, plant trays were kept in a plastic house suspended by their borders on metal rails 45 cm above the floor, with cell drainage holes unimpeded. Estimation of genetic parameters GI means and variances were calculated for each of the populations in each trial. Genetic (s2G), environmental (s2E) and phenotypic (s2P) variances, as well as broad sense heritability were calculated as indicated by Mather and Jinks (1977). Environmental variance (s2E) was estimated as the geometric mean of total variances in generations P1 and P2. Generation mean analyses were performed on P1, P2 and F2 (Mather and Jinks, 1977), to estimate the additive [a] and non-additive [d] mean components and the mean degree of dominance (MDD = [d] / [a]). Test of the hypothesis of monogenic inheritance Plant frequency distributions of GI were obtained for each generation studied in the trials. An arbitrary truncation point (TP) was chosen to permit a clear discrimination between the parental phenotypes: the TP value corresponded to a GI value below which fell most of the plants of the resistant parent, and above which fell most of the plants of the susceptible parent. Individual plant phenotypes were not classified into discrete classes (resistant/susceptible) because of the continuous nature of the variables under study. Data on the means and variances of P1 and P2 were used to calculate expected frequencies of plants below the TP in these populations, as well as in the F2, assuming monogenic inheritance and different presumed degrees of dominance. For each of the different presumed degrees of dominance, the monogenic hypothesis tested was based on the following suppositions and procedures: a) The GI follows a Poisson distribution. b) Parental generations P1 and P2 are assumed to have expected means (and variances) equal to their respective observed means. Frequencies of plants for which the GI value was below or equal to the truncation point (<TP) were obtained from the theoretical Poisson distribution assumed for P1 and P2, and were taken to represent the respective expected frequencies under the proposed model. c) The mean of the F1 population was taken as:
where: P1 and P2 are the respective parental means, and MDD is the mean degree of dominance under consideration. The variance of the F1 population was taken (under the Poisson distribution) as:
d) The frequency of F1 individuals with GI values <TP was estimated from the Poisson distribution assumed for that population, and was assumed to be the true (expected) frequency under the model. e) Under the hypothesis of monogenic inheritance, the expected frequency of plants in F2 with GI values <TP was calculated as the weighted average of the expected frequencies in P1, P2 and F1 (weights 1:2:1, respectively), as estimated in b and d. f) Expected numbers of plants with GI values <TP in P1, P2 and F2 were calculated by multiplying the expected frequencies (obtained in b, b and respectively) by the total number of plants tested per generation. g) Expected numbers of plants with values <TP in P1, P2 and F2 were compared with their respective observed values in each generation, and the significance of the deviations was estimated with a c2 test. The frequency of expected plants in P1 was added to that of P2, in order to avoid expected cell frequencies equal to zero. The number degrees of freedom in the c2 test was therefore 1. h) c2 Values significant at a = 0.05 would lead to the rejection of the hypothesis of monogenic inheritance under the degree of dominance considered. On the other hand, a nonsignificant value of c2 would not allow rejection of that hypothesis. Values of the c2 for each monogenic hypothesis/presumed MDD tested were plotted against their respective hypothetical MDD’s. The range of MDD values for which c2 values fell below the critical a = 0.05 value represented the MDD range for which the monogenic hypothesis was not rejected. RESULTS AND DISCUSSION The genetic variance estimate of nematode resistance measured by GI was much higher than the environmental variance, and broad-sense heritability was high (Table 1), indicating that GI is a trait subjected to a low degree of environmental influence. The MDD estimate was -0.3333, indicating a low degree of incomplete dominance in the direction of lower GI. This MDD point estimate, close to zero, indicates a predominantly (but not entirely) additive gene action of the gene(s) controlling nematode resistance - a result similar to that found by Gomes (1999) for the gene that controls resistance to M. incognita.
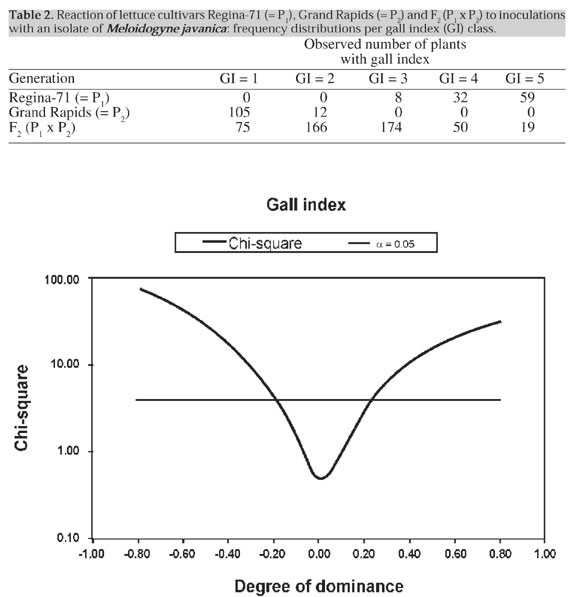
The high value obtained for the broad-sense heritabilities indicated that response to M. javanica in cultivar Grand Rapids is under the control of a single gene locus. However, the frequency distribution of GI in the F2 generation does not provide clear evidence for monogenic control (Table 2). Phenotypes could not be classified into the discrete classes of resistant or susceptible, due to the continuous nature of the variable under study. This property was associated with the predominantly additive gene action for the trait, which was responsible for the existence in F2 of GI’s intermediary between those of the parents. The proposed test of the monogenic hypothesis indicated nonetheless that frequency distributions of GI were consistent with segregation at a single locus. Within the degrees of dominance of -0.2 and -0.2 for segregation of a putative locus, the calculated c2 values fell below that of the a = 0.05 limit (Figure 1). These results strongly support the hypothesis of monogenic inheritance. The phenotypic distributions in the parental and F2 generations may therefore be explained by the action of a single locus with predominantly additive gene action.
The point estimate for the degree of dominance (Table 1) fell into a range close to the boundaries for degree of dominance within which the null hypothesis of monogenic control of resistance would not be rejected (Figure 1). These results provide evidence that a single gene locus, or at least a single chromosome segment, may be responsible for resistance to M. javanica in lettuce. In spite of the fact that there is clear evidence for one major locus controlling nematode resistance, the data do not preclude the existence of other genes (modifiers) with minor effects. Our conclusions are similar to those of Gomes (1999), who examined resistance to M. incognita in the same lettuce populations. The major allele involved in the M. javanica resistance of the lettuce cultivar Grand Rapids bears similarity of gene action to the Me allele responsible for resistance to M. incognita in the same cultivar. It is therefore possible that the Me allele itself could be responsible for resistance to both root nematode species. Our data, however, do not provide evidence to support or invalidate that conclusion. Consequently, the identity between the Grand Rapids alleles for resistance to both nematode species remains to be demonstrated or disproved in further trials. ACKNOWLEDGMENTS Research supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico/MCT)/ RHAE (Programa de Recursos Humanos em Áreas Estratégicas) grants to L.A.A. Gomes and A.C.B. Oliveira, and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais), CAPES/MEC, HortiAgro Sementes Ltda., and FAEPE/UFLA. We also thank the Instituto Agronômico do Paraná (IAPAR) and EMBRAPA/Hortaliças for providing isolates of Meloidogyne spp. REFERENCES Boneti, s.i.S. (1981). Inter-relacionamento de micronutrientes como parasitismo de Meloidogyne exigua em mudas de cafeeiro (Coffea arabica L.). M.S. Dissertation, Universidade Federal de Viçosa, Viçosa, MG, Brazil. Campos, V.P. (1995). Doenças causadas por nematóides em alcachofra, alface, chicória, morango e quiabo. Inf. Agropecu.17 (182): 17-22. Charchar, J.M. (1991). Comportamento de cultivares de alface à infecção por nematóides de galhas. Hortic. Bras. 9: 35 (Abstract). Charchar, J.M. and Moita, A.W. (1996). Reação de cultivares de alface à infecção por misturas populacionais de Meloidogyne incognita raça 1 e M. javanica em condições de campo. Hortic. Bras. 14: 185-189. Costa, C.P. and Silva, N. (1976). Melhoramento da alface para resistência múltipla ao calor e ao mosaico. Rev. Oleric.15: 26-27. Ferraz, S. and Mendes, M.L. (1992). O nematóide das galhas. Infor. Agropecu. 16 (172): 43-45. Gomes, L.A.A. (1999). Herança da resistência da alface (Lactuca sativa L.) cv. Grand Rapids ao nematóide de galhas Meloidogyne incognita (Kofoid and White) Chitwood. Doctoral thesis, Universidade Federal de Lavras, Lavras,MG, Brazil. Gomes, L.A.A., Mendes, W.P., Maluf, W.R., Azevedo, S.M., Freitas, J.A. and Moretto, P. (1996). Resistência de cultivares de alface à infecção por Meloidogyne incognita (raças 1, 2 e 3). Hortic. Bras. 14: 87 (Abstract). Hussey, R.S. and Barker, K.R. (1973). a comparison of methods collecting inocula of Meloidogyne spp. including a new technique. Plant Dis. Rep. 57: 1025-1028. Kaloo, D. (1988). Vegetable Breeding Vol. II. CRC Press, Boca Raton, FL, USA. Lordello, L.G.E. (1984). Nematóides das Plantas Cultivadas. Nobel, São Paulo, SP, Brazil. Maluf, W.R. (1997). Resistência a nematóides de galhas Meloidogyne spp. em espécies olerícolas. In: Resistência de Plantas a Doenças (Zambolin, L. and Ribeiro-do-Vale, F.X., eds.). Fitopatologia Brasileira/Congresso Brasileiro de Fitopatologia, 30, Palestras, pp. 57-63. Mather, K. and Jinks, J.L. (1977). Introduction to Biometrical Genetics. Cornell University Press, Ithaca, NY, USA. McGuire, P.E., Ryder, E.C., Michelmore, R.W., Clark, R.L., Antle, R., Emery, G., Hannan, R.M., Kesseli, R.V., Kurtz, E.A., Ochoa, O., Rubatzky, V.E. and Waycott, W. (1993). Genetic resources of lettuce and Lactuca species in California - an assessment of the USDA and UC collections and recommendations for long-term security. Genetic Resources Conservation Program, Division of Agriculture and Natural Resources, University of California, Davis, CA, USA, Report No. 12. Mendes, W.P. (1998). Hospedabilidade e resistência de cultivares de alface (Lactuca sativa L.) aos nematóides de galhas Meloidogyne incognita (raça 1,3 e 4) e Meloidogyne javanica. M.S. Dissertation, Universidade Federal de Lavras, Lavras, MG, Brazil. Nagai, H. (1979). Obtenção de novos cultivares de alface (Lactuca sativa L.) resistente ao mosaico e ao calor - Brasil 48, 202 e 221. Rev. Oleric. 17: 129-137. Nagai, H. (1980). Obtenção de novos cultivares de alface (Lactuca sativa L.) resistente ao mosaico e ao calor - Brasil-303 e 311. Rev. Oleric. 18: 14-21. Nagai, H. (1983). Caracterização de resistência ao calor em alface (Lactuca sativa L.). In: Congresso Brasileiro de Olericultura,23. Resumos. Sociedade de Olericultura do Brasil, Vitória, ES, Brazil. Nagai, H. and Costa, A.S. (1972). Resistência ao calor e ao mosaico numa variedade nova de alface tipo manteiga. Rev.Oleric. 12: 30 (Abstract). Nagai, H. and Costa, A.S. (1973). Seleção de novas linhagens de alface resistentes ao mosaico e ao calor. Rev. Oleric. 13: 27-28. Nagai, H. and Lisbão, R.S. (1980). Observação sobre resistência ao calor em alface (Lactuca sativa L.). Rev. Oleric. 18:7-13. Ryder, E.J. (1986). Lettuce breeding. In: Breeding Vegetable Crops (Basset, M. ed.). AVI Publishing Co., Westport, CT,USA, pp. 433-474. Silva, E.C., Maluf, W.R., Leal, N.R. and Gomes, L.A.A. (1999). Inheritance of bolting tendency in lettuce Lactuca sativa L. Euphytica 109: 1-7. |