ABSTRACT. Isolates of Bipolaris sorokiniana were analyzed by random-amplified polymorphic DNA (RAPD) techniques to determine the amount of intraspecific genetic variability and to study host-pathogen interactions. Ten isolates originated from different regions of Brazil were examined. Plants of the wheat cultivars BR8, BH1146 (original host) and IAC-5 Maringá, classified as resistant, moderately resistant or susceptible to B. sorokiniana, respectively, were inoculated with these 10 isolates. Twenty-seven isolates were recovered from these cultivars and were analyzed by RAPD assay and compared to the RAPD of the original 10 isolates. According to the RAPD profiles there was a high level of genetic variability among the isolates. We detected 69 polymorphic fragments, ranging from 1.6 to 0.54 kb, in the original 10 isolates; 57 fragments with sizes between 1.98 and 0.38 kb from the isolates recovered from BH1146; 47 polymorphic bands, ranging from 1.96-0.54 kb, were detected in the isolates from BR8 and 32 fragments between 1.98 and 0.42 kb in isolates were recovered from IAC-5 Maringá. The number of polymorphic fragments varied, even for the same isolate, when the isolates were recovered from different cultivar hosts. Key words: Bipolaris sorokiniana, Wheat, Genetic variability, RAPD INTRODUCTION One of the many diseases affecting wheat crops in Brazil is spot blotch, caused by Bipolaris sorokiniana (Sacc. in Sorok) Shoem, anamorphic state, syn. Helminthosporium sativum Pamm. King and Bakke, syn. Drechslera sorokiniana (Sacc.) Subram & Jaim, telomorphic state Cochliobolus sorokiniana (Ito & Kurib) Drechsl. ex Daxtur. Bipolaris sorokiniana infects plant leaves, stems, crowns and roots, causing infected tissues to become necrotic. This pathogen is not specialized, as it affects wheat, triticale, barley and most grasses (Agrios, 1997). Historically, B. sorokiniana has been described as a variable fungus with many morphological (Christensen, 1925; Christensen and Davis, 1937; Tinline, 1960; Matsumura, 1991; Valim-Labres, 1995; Oliveira et al., 1998) and physiological variants (Tinline, 1960; Wood, 1962; Nelson and Klyne, 1962; Hart, 1979; Metha, 1981; Valim-Labres et al., 1997). Part of this variability has been attributed to heterokaryosis and parasexuality mechanisms (Tinline, 1962). Characterization of this fungus is fundamental for the study of host-parasite interactions, in order to develop appropriate strategies for plant breeding programs. Mendelian genetic analysis has been decisive for the discovery of many fundamental principles in plant pathology. However, important phenomena observed in natural populations of fungi are not adequately explained by Mendelian inheritance of phenotypic characters. These include an unusually high mutation rate, the absence of detectable reversion and a frequent association of mutation with other phenotypic changes, including loss of pathogenicity (Bronson et al., 1990; Chumley and Valent, 1990; Levi et al., 1991; Klister and Miao, 1992). Analyses of the diversity of plant pathogens and studies of the interaction with their hosts have been revolutionized by molecular techniques. Particularly, the polymerase chain reaction (PCR) assay has provided a framework to understand taxonomy and population structure. The assessment of genetic diversity is required for the development of long-time disease management strategies. The random-amplified polymorphic DNA (RAPD) method has been successfully used to identify strains (Guzmán et al., 1999; Chiocchetti et al., 1999; Pryor and Gilbertson, 2000), to characterize races (Malvick and Grau, 2001) and to analyze virulence variability related to genetic polymorphisms (Chen et al., 1995; Chen et al., 1999; Browning et al., 1999; Chakraborty et al., 1999; Kolmer and Liu, 2000) in phytopathogenic fungi. It has also been used in the study of inter- and intraspecific variability among populations from different (Vakalounakis and Fragkiadakis, 1999; Pimentel et al., 2000; Pažoutová et al., 2000) and from the same geographic regions (Bhat and Subbarao, 1999; Walker et al., 2001; Ma et al., 2001). Bipolaris sorokiniana has a high degree of phenotypic variability; however, the genetic diversity of this fungus has not been fully studied. Studies have been made of isozyme polymorphisms among isolates (Matsumura, 1991; Valim-Labres, 1995). However, no such work has been done using DNA polymorphisms. We used a RAPD assay to determine the genetic variability of B. sorokiniana isolates collected from different regions of the country and examined the host-pathogen interactions by analyzing RAPD polymorphisms in isolates of this fungus recovered from wheat cultivars with different resistance levels. MATERIAL AND METHODS Fungal Isolates We used 10 isolates from different geographic regions of Brazil, provided by the Centro Nacional de Pesquisa do Trigo, EMBRAPA-CNPT (Table 1). These isolates were inoculated on the wheat cultivars BR8, BH1146 (original host) and IAC-5 Maringá, which are classified as resistant, moderately resistant and susceptible to B. sorokiniana infection, respectively (Oliveira et al., 1998). After the fungus had been re-isolated from the wheat cultivars, 27 new samples were collected and were also analyzed in this study.
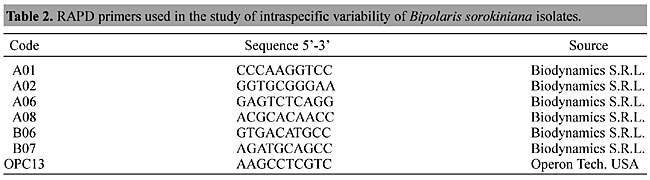
DNA isolation Fungal cultures were grown on potato-dextrose-agar (PDA) incubated at 24 ± 2°C on a 12-h light/12-h dark cycle for seven days. Aliquots of the cultures were used to inoculate flasks containing 50 ml of potato-dextrose broth, which were incubated as described above. Twenty grams of mycelium was used for DNA isolation according to Ashktorab and Cohen (1992), with some modifications. The mycelia were collected by filtration and the samples ground to a fine powder in liquid nitrogen. One volume (w/v) of lysis buffer (200 mmol/l Tris-HCl, pH 8.0; 250 mmol/l NaCl, 25 mmol/l EDTA, pH 8,0; 1% sodium dodecyl sulfate) with 1% ß-mercaptoethanol and 50 µg/ml proteinase K (Gibco-BRL) was added to the powder. The samples were then incubated at 65°C for 1 h. After incubation, samples were centrifuged for 20 min at 13,000 g. The supernatant was collected and one extraction with phenol was performed, followed by two extractions with phenol-chloroform (1:1) and one with chloroform ”. Fifty micrograms RNAse A (Sigma Aldrich) was added to the samples and incubated at 37°C for 15 min. The DNA was precipitated with 0.1 vol. of sodium acetate 3 mol/l and 2.5 vol. of isopropanol. The DNA was collected carefully and washed with 70% ethanol, dried and then dissolved in TE (10 mmol/l Tris-HCl, 1 mmol/l EDTA). RAPD analysis The primers used in this study were A1-A10 and B1-B10 (Biodynamics Kits S.R.L.), OP13, OPB17 and OPB03 (Operon Technologies Inc., USA). For standardization of the amplification conditions and selection of the primers, the genomic DNA extracted from original samples of B. sorokiniana was used as a template for the amplifications. The primers A01, A02, A06, A08, B06, B07 and OPC13 (Table 2) were chosen due to the consistent reproducibility of their amplification products.
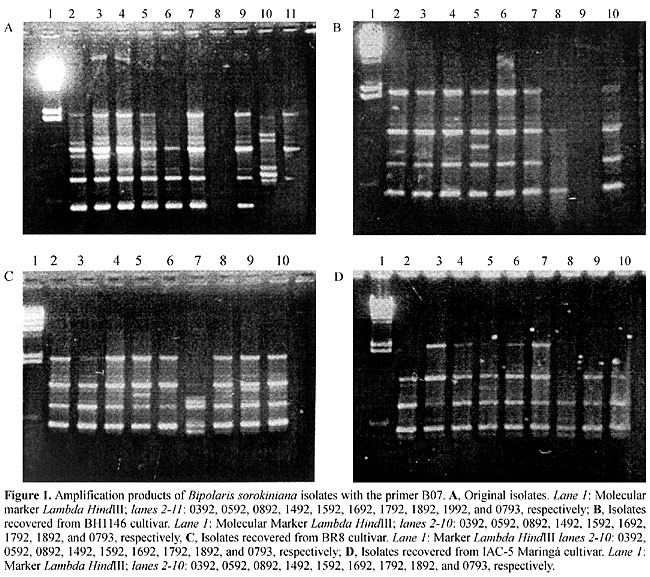
Amplification conditions were slightly modified from those indicated in Williams et al. (1990). All reactions were carried out in a volume of 25 µl with 30 ng DNA; 2.5 mmol/l of each dNTP (dATP, dGTP, dCTP, dTTP); 60 ng of primer; 2.5 µl of 10X reaction buffer (10 mmol/l Tris-HCl, pH 8.0, 2.0 mmol/l MgCl2, 0.01% gelatine); 50 µg/ml bovine serum albumin (Gibco-BRL), and 1 unit Taq DNA polymerase (CENBIOT/RS-BR). The reactions were performed on a thermal cycler (MJ Research) programmed for one initial cycle with 1 min at 94°C, 5 min at 35°C; 2 min at 72°C and 45 cycles of 1 min at 94°C, 1 min at 35°C; 2 min at 72°C. The assays were repeated at least twice with each primer in different experiments. Amplification products were electrophoresed in 1.4% agarose gel, and detected by staining with ethidium bromide. Data analysis The RAPD data were analyzed using the Statistical Package for the Social Sciences (SPSS) software, 2nd edition (Wilkinson et al., 1992), through which similarity coefficients were calculated and a dendrogram for genetic distances constructed. Similarity was evaluated through simple association and the genetic distance determined as the Euclidian Distance. The binary matrix was built pair-wise, and the presence or absence of a determined RAPD band scored 1 and 0, respectively. The hierarchical groupings were based on the unweighted pair set method using average (UPGMA) (Crisci and Armengol, 1983). RESULTS In this investigation the genetic variation existing among isolates of B. sorokiniana and the host-pathogen interactions was studied by analyzing the DNA polymorphisms in isolates recovered from wheat cultivars with different resistance levels to B. sorokiniana infection. The RAPD profiles showed a high level of genetic variability among the B. sorokiniana isolates. Variability was observed among isolates originated from the same cultivar, as well as between different host cultivars. Sixty-nine polymorphic fragments were detected in the 10 original samples (BH1146 host), ranging from 1.6 to 0.54 kb. Polymorphisms, were also observed among the isolates recovered from the BH1146 cultivar, in which 57 polymorphic fragments were identified, some of which had not been observed in the original samples (Figure 1A and B). The amplifications with all seven primers generated fragments ranging from 1.98 to 0.38 kb.
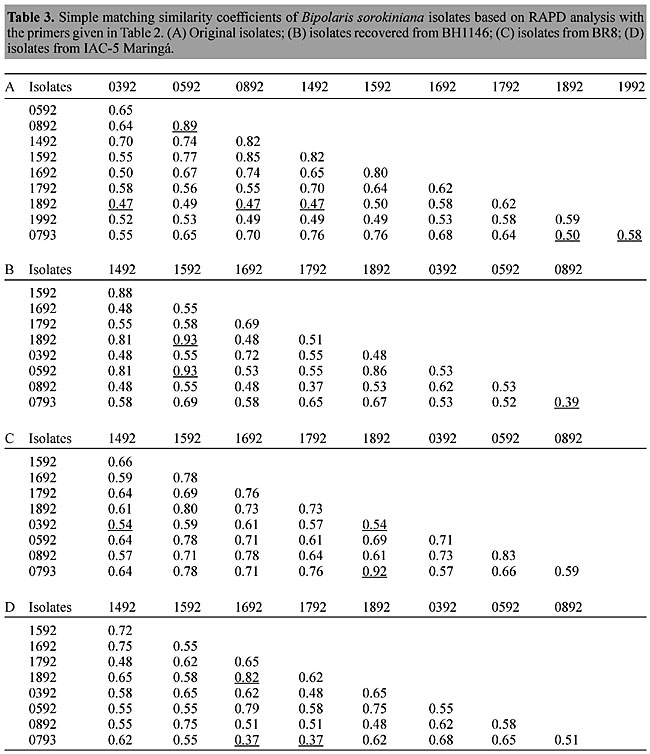
The amplification products of the isolates recovered from cultivars BR8 and IAC-5 Maringá also showed genetic variability (Figure 1). Forty-seven polymorphic fragments ranging from 1.96-0.54 kb were identified in isolates from cultivar BR8, and 32 polymorphic fragments were scored, ranging from 1.98-0.42 kb, in isolates from cultivar IAC-5 Maringá. Some isolates did not amplify with this primer (B07), even after many repetitions. Amplification problems are sometimes due to low quality of the DNA template. However, this possibility was ruled out because these same samples amplified with other primers. Computer analysis of these data resulted in a matrix with similarity coefficients for each set of isolates (Table 3). Considerable polymorphism was found, with a similarity index below 50%. The highest similarity coefficient among the original isolates was 0.89 for isolates 0892 and 0592, and the lowest coefficient 0.47 was seen for the isolate 1892 in relation to isolates 0392, 0892 and 1492. With the isolates recovered from cultivar BH1146 the maximum coefficient was 0.93 for isolate 1592 in relation to 0592 and 1892 isolates, and the lowest coefficient 0.39 was observed for pair 0793-0892; for the isolates from BR8 the highest value was 0.92 for isolates 0392 and 1492, and 0.54 (the lowest) for isolate 0392 in relation to 0793 and 1892, and for those from IAC-5 Maringá the results were 0.82 for pair 1892-1692, and the lowest coefficient 0.37 for isolate 0793 with 1692 and 1792.
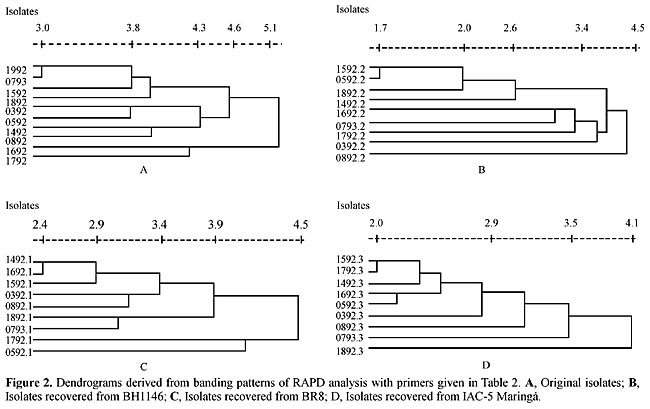
The dendrograms were constructed with the RAPD data generated by the amplified fragments from the original isolates recovered from BR*, BH1146 and IAC-5 Maringá cultivars, respectively (Figure 2). There was genetic variation among the isolates because the isolates never appeared in the same group. Also, the groupings showed no association with the geographic origin of the isolates.
DISCUSSION According to Burdon and Silk (1997), plant pathogenic fungi most commonly rely on mutation and recombination as the main source of genetically based variation. Within a species, gene flow between populations supplements these processes as propagules spread from one epidemiological area to another, and from one deme to the next. Gene flow, along with other evolutionary forces, can result in the spread of single genes (or DNA sequences), genotypes, and even in the establishment of whole populations in different regions (McDermott and McDonald, 1993). Based on the high degree of genetic similarity found among B. sorokiniana isolates from Passo Fundo, RS, Dourados, MS and Planaltina, GO, we suggest that there has been considerable gene flow among all these populations. However, more isolates from each region and from different regions must be tested with a higher number of primers in order to confirm this hypothesis. Much research with B. sorokiniana has been done during the past years, most of it addressing changes in colony characteristics and in virulence in isolates recovered from successive infections of the plant host (Christensen and Devay, 1955; Kinsey and Hooker, 1973; Castro, 1976; Linden, 1989). We observed modifications in the number of amplified fragments of fungal isolates from wheat cultivars with different resistance levels to B. sorokiniana infection. It is well known that the plant host exerts a strong selective pressure on its pathogens (Burdon and Silk, 1997). Nevertheless, in this investigation the relationship between the host resistance to infection and DNA polymorphisms was not established as some of the fragments that amplified with the isolates recovered from cultivar BH1146 did not amplify with the original isolates that came from the same cultivar. These isolates had been previously analyzed for their morphological and cultural characteristics on PDA medium, as well as virulence for wheat cultivars BH1146, BR8 and IAC-5 Maringá (Oliveira et al., 1998). Different degrees of virulence and high morphological, as well as growth rate variability were found. However, no association was observed between morphological and cultural characteristics, virulence and RAPD patterns. ACKNOWLEDGMENTS The authors thank CNPq, RHAE and CAPES for their support of this research. REFERENCES Agrios, G.N. (1997). Plant Pathology. 4th edn. Academic Press, San Diego, CA, USA. Ashktorab, H. and Cohen, R.J. (1992). Facile isolation of genomic DNA from filamentous fungi. BioTechnics 13: 198-200. Bhat, R.G. and Subbarao, K.V. (1999). Host range specificity in Verticillium dahliae. Phytopathology 89: 1218-1225. Bronson, C.R., Taga, M. and Yoder, O.C. (1990). Genetic control and distant segregation of T-toxin production in field isolates of Cochliobolus heterostropus. Phytopathology 80: 819-823. Browning, M., Rowlwy, L.V., Zeng, P., Chandlee, J.M. and Jackson, N. (1999). Morphological, pathogenic and genetic comparisons of Colletotrichum graminicola isolates from Poaceae. Plant Dis. 83: 286-292. Burdon, J.J. and Silk, J. (1997). Sources and patterns of diversity in plant pathogenic fungi. Phytopathology 87: 664-669. Castro, C. (1976). Mudanças em esporulação e patogenicidade de Helminthosporium sativum Pamm. King & Bakke através de passagens seriadas em hospedeiros. MSc. thesis, Escola Superior de Agricultura Luiz de Queiroz, Piracicaba, SP, Brazil. Chakraborty, S., Perrott, R., Ellis, N. and Thomas, M.R. (1999). New aggressive Colletotrichum gloeosporioides strains on Stylosanthes scabra detected by virulence and DNA analysis. Plant Dis. 83: 333-340. Chen, X., Line, R.F. and Leung, H. (1995). Relationship between virulence variation and DNA polymorphism in Puccinia striformis. Phytopathology 83: 1489-1497. Chen, X., Romaine, C.P., Tan, Q., Schlagnhaufer, B., Ospina-Giraldo, M.D., Royse, D.J. and Huff, D.R. (1999). PCR-based genotyping of epidemic and preepidemic Trichoderma isolates associated with green mold of Agaricus bisporus. Appl. Environ. Microbiol. 65: 2674-2678 Chiocchetti, A., Ghignone, S., Minuto, A., Gullino, M.L., Garibaldi, A. and Migheli, Q. (1999). Identification of Fusarium oxysporum f.sp. basilici isolated from soil, basil seed, and plants by RAPD analysis. Plant Dis. 83: 576-581. Christensen, J.J. (1925). Physiology specialization and mutation in Helminthosporium sativum. Phytopathology 15: 785-795. Christensen, J.J. and Davis, F.R. (1937). Nature variation in Helminthosporium. Mycologia 29: 85-99. Christensen, J.J. and Devay, J.E. (1955). Adaptation of plant pathogen to host. Annu. Rev. Plant Physiol. 6: 367-392. Chumley, F.G. and Valent, B. (1990). Genetic analysis of melanin-deficient nonpathogenic mutants of Magnaporthe grisea. Mol. Plant Microbe Interact. 3: 135-143. Crisci, J.V. and Armengol, F.L. (1983). Introduction a la Teoria Practica de la Taxonomia Numerica. Secretaria General de la Organización de los Estados Americanos, Washington, DC, USA. Guzmán, P., Gepts, P., Temple, S., Mkandawire, A.B.C. and Gilbertson, R.L. (1999). Detection and differentiation of Phaeoisariopsis griseola isolates with the polymerase chain reaction and group-specific primers. Plant Dis. 83: 37-42. Hart, G.E. (1979). Evidence for a triplicate set of glucose phosphate isomerase structural genes in hexaploid wheat. Biochem. Genet. 17: 585-598. Kinsey, J.G. and Hooker, A.L. (1973). Changes in Helminthosporium turcicum populations following seried host passage. Plant Dis. Rep. 57: 590-591. Klister, C.H. and Miao, V.P.W. (1992). New modes of genetic change in filamentous fungi. Annu. Rev. Phytopathol. 30: 131-152. Kolmer, J.A. and Liu, J.Q. (2000). Virulence and molecular polymorphism in international collections of the wheat leaf rust fungus Puccinia triticina. Phytopathology 90: 427-436. Levi, M., Romao, J., Marchetti, M.A. and Hamer, J.E. (1991). DNA fingerprinting with a dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell. 3: 95-102. Linden, A.R. (1989). Alterações na patogenicidade, morfologia e atividade isoesterásica de Helminthosporium sativum decorrentes de passagens sucessivas por duas cultivares hospedeiras de trigo. B.Sc. thesis in Biological Sciences, Departamento de Genética, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil. Ma, Z., Boehm, E.W.A., Luo, Y. and Michailides, T.J. (2001). Population structure of Botryosphaeria dothidea from pistachio and other hosts in California. Phytopathology 91: 665-672. Malvick, D.K. and Grau, C.R. (2001). Characteristics and frequency of Aphanomyces euteiches races 1 and 2 associated with alfalfa in the midwestern United States. Plant Dis. 85: 740-744. Matsumura, A.T. (1991). Variabilidade intraespecífica quanto à patogenicidade, características da cultura e padrão isoesterásico em populações naturais de Bipolaris sorokiniana (Helminthosporium sativum). Ph.D. thesis, Departamento de Genética, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil. McDermott, J.M. and McDonald, B.A. (1993). Gene flow in plant pathosystems. Annu. Rev. Phytopathol. 31: 353-373. Metha, Y.R. (1981). Identification of races of Helminthosporium sativum of wheat in Brazil. Pesqui. Agropecu. Bras. 16: 331-336. Nelson, R.R. and Klyne, D.M. (1962). Intraspecific variation in pathogenicity in the genus Helminthosporium to gramineous species. Phytopathology 52: 1045-1049. Oliveira, A.M.R., Matsumura, A.T.S., Prestes, A.M., Matos, G.S. and Van Der Sand, S.T. (1998). Variabilidade patogênica e morfológica em isolados de Bipolaris sorokiniana. Fitopatol. Bras. 23: 349-353. Pažoutová, S., Bandyopadhyay, R., Frederickson, D.E., Mantle, P.G. and Frederickson, R.A. (2000). Relation among sorghum ergot isolates from the Americas, Africa, India and Australia. Plant Dis. 84: 437-442. Pimentel, G., Preever, T.L. and Carris, L.M. (2000). Genetic variation among natural populations of Tilletia controversa and T. bromi. Phytopathology 90: 376-383. Pryor, B.M. and Gilbertson, R.L. (2000). A PCR-based assay for detection of Alternaria radicina on carrot seed. Plant Dis. 85: 18-23. Tinline, R.D. (1960). Pathogenic and cultural variation in Cochliobolus sativus. Trans. Br. Mycolol. Soc. 43: 696-697. Tinline, R.D. (1962). Cochliobolus sativus. V. Heterocariosis and parasexuality. Can. J. Bot. 40: 425-437. Vakalounakis, D.J. and Fragkiadakis, G.A. (1999). Genetic diversity in Fusarium oxysporum isolates from cucumber: Differentiation by pathogenicity, vegetative compatibility, and RAPD fingerprinting. Phytopathology 89: 161-168. Valim-Labres, M.E. (1995). Variabilidade intra-específica em Bipolaris sorokiniana: análise morfológica, isoenzimática e de patogenicidade. M.Sc. thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil. Valim-Labres, M.E., Porto, M.D. and Matsumura, A.T.S. (1997). Effects of host resistance on the isozymatic patterns of Bipolaris sorokiniana (Dematiaceae, Moniliales). Braz. J. Genet. 20: 541-545. Walker, S.L., Leath, S., Hagler Jr., W.M. and Murphy, J.P. (2001). Variation among isolates of Fusarium graminearum associated with Fusarium head blight in North Carolina. Plant Dis. 85: 404-410. Wilkinson, L.J., Hill, M., Welna, J.P. and Birkenbeuel, G.K. (1992). Statistics. Systat Inc., Chicago, IL, USA. Williams, J.G.K., Kubelik, A.R., Kennethj, L., Rafalski, J.R. and Tingey, S.V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18: 6231-6235. Wood, L.S. (1962). Relation of variation in Helminthosporium sativum to seedling blight of small grains. Phytopathology 52: 493-497. |
|