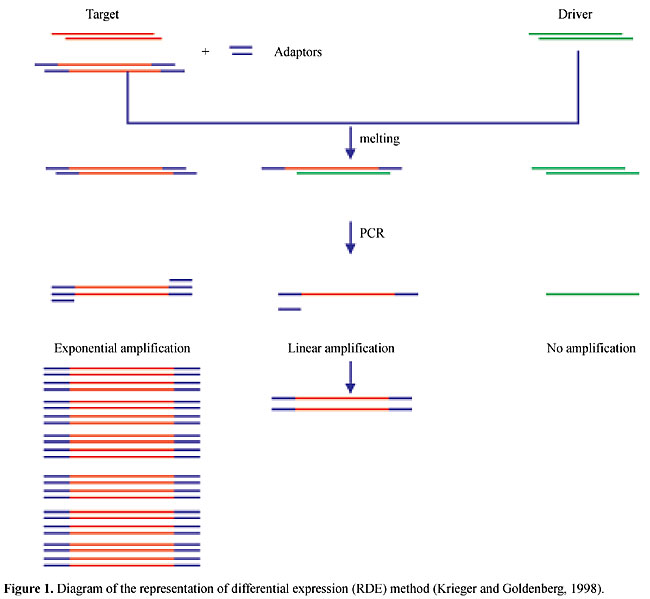
ABSTRACT. The process of Trypanosoma cruzi metacyclogenesis involves the transformation of noninfective epimastigotes into metacyclic trypomastigotes, which are the pathogenic form. The analysis of stage-specific genes during T. cruzi metacyclogenesis may provide insight into the mechanisms involved in the regulation of gene expression in trypanosomatids. It may also improve the understanding of the mechanisms responsible for the pathology of Chagas disease, and could lead to the identification of new targets for chemotherapy of this disease. We have demonstrated that during metacyclogenesis the expression of several genes is controlled at the translational level by an alternative regulatory mechanism. This mechanism may involve the mobilization of mRNA to the translation machinery. We have been using self-made T. cruzi microarrays to investigate the role of polysomal mobilization in modulating gene expression during metacyclogenesis. Key words: Trypanosoma cruzi, Metacyclogenesis, Stage-specific gene THE LIFE CYCLE OF TRYPANOSOMA CRUZI The trypanosomatid Trypanosoma cruzi is the causative agent of Chagas disease (Chagas, 1909), a parasitic disease affecting more than 20 million individuals in Latin America. For this reason, T. cruzi is one of the most important parasitic models and is studied by various research groups throughout the world. The relevance of studies on the biology of this parasite also comes from the fact that T. cruzi belongs to an evolutionary primitive eukaryotic group. The life cycle of this parasite involves two intermediate hosts (triatomine insects and mammals, including man) and three well-defined morphological and functional developmental stages: epimastigotes, trypomastigotes and amastigotes (de Souza, 1984). The epimastigote forms replicate in the midgut of the insect host and develop into nonreplicative metacyclic trypomastigote forms. When the insects feed on blood, they release in their excreta metacyclic trypomastigotes that penetrate the body of the mammalian host through the wound and are able to invade cells. Within the host cells, the parasite differentiates into the replicative amastigote form. After multiplication, the amastigotes differentiate into blood stream trypomastigotes that are released into the circulatory system, infecting new cells. THE METACYCLOGENESIS PROCESS Metacyclogenesis is the process by which noninfective epimastigotes are transformed into metacyclic trypomastigotes, the pathogenic form. Understanding this process may pave the way towards the development of new treatments to control this disease. During in vivo metacyclogenesis, differentiating epimastigotes adhere to the epithelium of the insect rectum (Boker and Shaub, 1984) before transformation into metacyclic trypomastigotes (de Souza, 1984). Epimastigotes colonize the insect’s midgut and rectum, whereas trypomastigotes are found mainly in the rectum, weakly attached to the rectal cuticle (Schaub, 1989; Kollien et al., 1998). Epimastigotes adhere to the epithelium via the tip of the flagellum, and indirectly by interdigitations with other parasites (Kollien et al., 1998). Adhesion is thought to be a prerequisite for differentiation into the infectious stage and it involves unspecific hydrophobic interactions with the superficial cuticle layer of the gut (Böker and Schaub, 1984; Zeledon et al., 1984). Different mechanisms and target molecules have been implicated in the adhesion of trypomastigotes to the insect epithelium. It has been suggested that chitin, chitosan and heparin are natural substrates for the binding of flagellate parasites (Wallbanks et al., 1989). In the rectal epithelium, chitin is present mainly in the procuticle layer, with tiny amounts in the superficial layer (Schmidt et al., 1998). Chitin is therefore not accessible to the parasites, as they colonize the intact intestinal wall, unless they can penetrate or disrupt the cuticle layer (Kollien et al., 1998). It is not clear how adhesion triggers the differentiation process but it has been demonstrated that both adhesion and metacyclogenesis are triggered by nutritional stress (Figueiredo et al., 2000). Moreover, previous work has shown that T. cruzi metacyclogenesis is stimulated by cyclic AMP and adenylate cyclase activators (Gonzales-Perdomo et al., 1988; Fraidenraich et al., 1993; Garcia et al., 1995). Although several factors have been already identified, the mechanisms and events involved with the onset of metacyclogenesis remain to be elucidated. However, it is known that the processes involved with the morphological changes that occur during T. cruzi differentiation are related to the expression of several stage-specific genes (Heath et al., 1990; Cooper et al., 1991; Bonaldo et al., 1991; Schenkmam et al., 1994; Di Nóia et al., 1995; Teixeira et al., 1995; Tomás and Kelly, 1996; Tomás et al., 1997). The metacyclogenesis process is a suitable model system for studying T. cruzi differentiation because this process can be mimicked in vitro, and conditions have been established resulting in the production of metacyclic trypomastigotes (Contreras et al., 1985). Briefly, this is accomplished by submitting epimastigote cells to nutritional stress in triatomine artificial urine (TAU) medium and further incubation in TAU supplemented with amino acids and glucose (TAU3AAG). In vitro metacyclogenesis promotes a metabolic synchronization of the cells; hence parasites at various stages of metacyclogenesis can be harvested from culture and their biological properties and gene expression products investigated. This greatly facilitates studies on differential gene expression, on the molecular mechanisms of morphogenetic regulation, on the properties of stage-specific antigens and on metabolic pathways. STAGE-SPECIFIC GENE EXPRESSION DURING METACYCLOGENESIS The analysis of stage-specific genes during T. cruzi metacyclogenesis may provide new insight into the mechanisms involved in the regulation of gene expression in trypanosomatids. It may also improve the understanding of the mechanisms responsible for the pathology of Chagas disease and may lead to the identification of new targets for chemotherapy of this disease. At the molecular level, very little is known about the process of metacyclogenesis and the changes that occur in the stage-regulated gene expression program. Previous work demonstrated that stage-specific gene expression precedes the morphological changes that occur in the differentiation of epimastigotes into metacyclic trypomastigotes (Contreras et al., 1985a; Goldenberg et al., 1985). The main differences in the polypeptide profile during in vitro differentiation of T. cruzi Dm28c are observed within the first 24 h of differentiation; after 12 h of incubation in differentiation medium, epimastigotes start to express metacyclic-specific antigens (Contreras et al., 1985a). After 6 h some differentiating epimastigotes are resistent to lysis mediated by complement, which is characteristic of trypomastigote forms (Sher et al., 1983). Changes in the lipid composition of the membranes of the parasite precede the morphological transformation of epimastigotes into metacyclic trypomastigotes (Esteves et al., 1989). Similar changes are observed in the carbohydrate composition at the surface of the parasites (Andrade et al., 1991). All these findings support the hypothesis that biochemical and gene expression changes precede morphological transformations of T. cruzi during the differentiation process. Relatively few T. cruzi stage-specific genes have been described so far, and most of them encode major surface antigens (Schenckman et al., 1994; Teixeira et al., 1995; Tomás and Kelly, 1996). The identification and characterization of stage-specific genes should improve our knowledge about T. cruzi differentiation and the metacyclogenesis process in particular. For this reason, we are interested in identifying and characterizing genes that are differentially expressed during the metacyclogenesis process. We have developed a method for isolating trypanosomatid stage-specific genes (Krieger and Goldenberg, 1998). This method, named representation of differential expression (RDE), consists of the selective amplification of genes specifically expressed by a given population (tester), followed by several rounds of subtraction with a reference population (driver) (Figure 1).
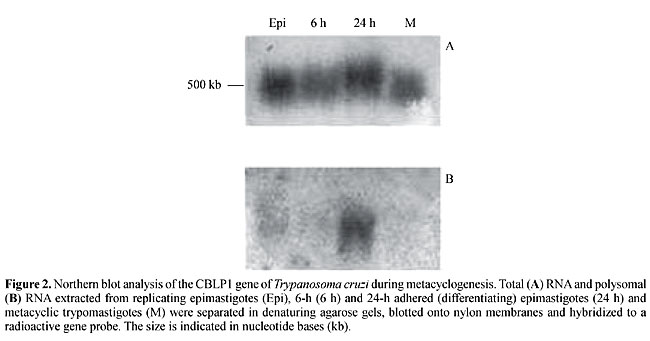
Several genes differentially expressed during T. cruzi metacyclogenesis have been cloned by using this method (Krieger et al., 1999). Northern blot analysis of RDE genes showed that some of them are transiently expressed during the in vitro metacyclogenesis process, while others are expressed only by differentiating cells and metacyclic trypomastigotes. Most of the nucleotide sequences of these RDE clones have shown no significant homology with sequences available in gene databases. We are currently interested in identifying the biological function of these genes. We have recently characterized two stage-specific genes, isolated from metacyclic trypomastigotes, with the RDE procedure (Yamada-Ogatta, S.F., Toma, H.K., Monteiro-Goes, V., Ávila, A.R., Dallagiovana, B.M., Goldenberg, S. and Krieger, M.A., unpublished results). These genes were named metacyclin I and II (Met-I and Met-II). They are present as single copy genes in the T. cruzi Dm28c genome. Northern and Western blot analyses showed that Met-I and Met-II are expressed by trypomastigotes, but not by epimastigotes. Analysis of the deduced amino acid sequences of Met-I and Met-II showed no particular features. Met-I encodes a basic polypeptide (21,888 kDa), whereas Met-II encodes an acidic polypeptide (22,261 kDa). The Met-I protein has three nuclear localization signals (Dingwall and Laskey, 1991). It also has four potential phosphorylation sites for protein kinase C and cAMP-dependent protein kinase. Nevertheless, further analysis is necessary to determine whether or not Met-I is involved with a signal transduction pathway in the metabolism of metacyclic trypomastigotes. Additional studies on the overexpression or inhibition of Met-I and Met-II would help determine the biological function of these proteins in metacyclic trypomastigotes. We also characterized a multicopy gene family differentially expressed during meta-cyclogenesis (Dallagiovanna et al., 2001). The genes of this family were isolated by RDE, and they encode polypeptides with chitin-binding domains, with eight similarly positioned half-cysteines and conserved aromatic residues involved in chitin recognition. The mRNAs for the members of this gene family were present in the total RNA fraction but were differentially mobilized to the polysomal fraction of adhered (differentiating) epimastigotes during meta-cyclogenesis, with a peak of accumulation at 24 h of differentiation (Figure 2). Polyclonal antiserum against the recombinant protein was used to analyze the synthesis of the proteins of this family during metacyclogenesis. Western blot analysis of T. cruzi extracts, using the specific antiserum, detected small 7- and 11-kDa polypeptides. The 11-kDa protein was present in similar amounts in the various cell populations, whereas the 7-kDa protein displayed differential expression during metacyclogenesis, with maximal levels in 24-h adhered (differentiating) epimastigotes. Further biochemical analysis is necessary to determine whether members of this protein family are involved in the adhesion of differentiating epimastigotes to the substrate during the meta-cyclogenesis process.
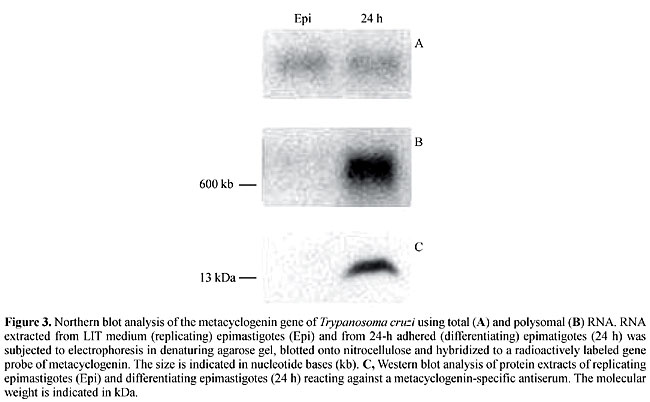
We have also described another gene isolated with RDE, which is specifically expressed by 24-h adhered (differentiating) epimastigotes (Ávila et al., 2001). This gene encodes a protein (metacyclogenin), with a molecular weight of 13 kDa, present only in differentiating epimastigotes. Northern blot analysis showed that this gene encodes a 630-nucleotide mRNA present in the total RNA extracted from both replicating epimastigotes and differentiating epimastigotes. However, when the analysis was carried out using polysomal RNA, the mRNA was detected only in differentiating epimastigotes (Figure 3). Consequently, we suggest that the expression of metacyclogenin is post-transcriptionally regulated and involves the mobilization of mRNA to polysomes (polysomal mobilization) during differentiation. No similarity has been found for the nucleotide sequences of this gene with sequences in gene databases, and the biological function of its product remains unknown. We are currently investigating the role of this gene in T. cruzi metacyclogenesis.
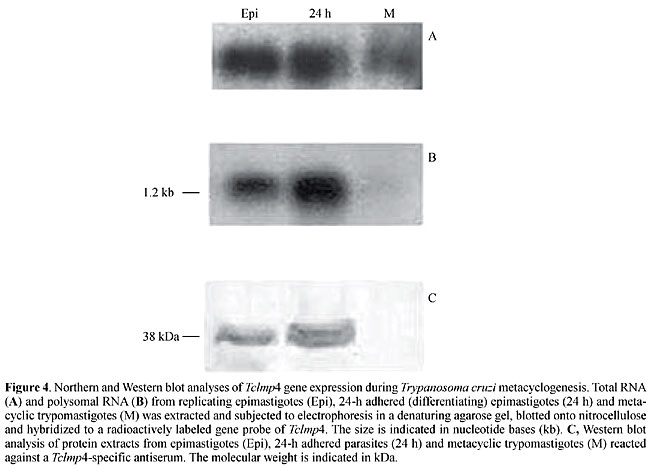
Another gene that was also cloned with the RDE method, which was characterized in our laboratory, appears to be regulated by polysomal mobilization (Fragoso et al., 2003). In this case, the gene, named Tclmp4, encodes a putative protein associated with U3, a small nucleolar ribonucleoprotein in T. cruzi. A 1.2-kb transcript was detected in polysomal RNA from epimastigotes and from differentiating parasites, but not in metacyclic trypomastigote forms. However, the intensity of the signal for this transcript was quite similar in the total RNA extracted from three different T. cruzi forms (Figure 4). Similar results were observed with the gene encoding topoisomerase II (Topo II, Fragoso et al., 1998). Topo II is not expressed in metacyclic trypomastigotes, although similar levels of mRNA are observed in all developmental stages of the parasite. Based on these observations, the transcript apparently is stored in the cytoplasm in a translation-repressed form in metacyclic trypomastigote forms.
This mechanism of translation regulation could be common to other trypanosomatid genes. In the case of T. cruzi cruzipain, the steady-state level of mRNA is similar in total RNA from all stages in the life cycle, although protein levels are five times higher in epimastigotes than in amastigotes or in trypomastigotes (Tomás and Kelly, 1996). The mRNA of the T. brucei Nrk protein gene is constitutively synthesized but is translated almost exclusively in stumpy trypomastigote blood forms (Gale et al., 1994). Gene expression in trypanosomatids is regulated essentially at the post-transcriptional level (Nozaki and Cross, 1995; Vanhamme and Pays, 1995; Teixeira, 1998) and this regulation occurs at the level of primary transcript processing (trans-splicing and polyadenylation), mRNA stability or translation product stability (Vanhamme and Pays, 1995). Previous work has provided evidence that T. cruzi mRNAs is stored in the cytoplasm in a translation-repressed form (Goldenberg et al., 1985), suggesting that mRNA mobilization to the polysomes is a post-transcriptional mechanism of gene expression regulation in trypanosomatids. Based on the results discussed above, it appears that during T. cruzi differentiation some transcripts are either mobilized to polysomes or stored in a compartment in which translation does not occur, being available for translation whenever necessary. Most studies of gene expression regulation in trypanosomatids involve genes abundantly expressed at a given development stage, such as the genes encoding variant surface glycoprotein, procyclin (Pays and Nolan, 1998), metacyclic variant surface glycoprotein (Graham et al., 1998), 85-kDa glycoprotein (Abuin et al., 1999) and amastin (Teixeira et al., 1995). Translation regulation may be involved, but other mechanisms of gene expression regulation appear to predominate. Our strategy of using polysomal RNA for the RDE procedure and analysis of different stages in metacyclogenesis allowed us to determine an alternative mechanism for gene expression regulation during the differentiation of T. cruzi, probably involving polysomal mobilization of expressed genes. The mechanism and the components that are involved in polysomal mobilization remain to be determined, as well as whether this is a general mechanism or is restricted to particular sets of genes. PERSPECTIVES During metacyclogenesis the expression of several genes is controlled at the translational level by an alternative regulation mechanism consisting of the mobilization of mRNAs to the translation machinery. Untranslated regions of kinetoplastid mRNA molecules are involved in post-transcriptional regulation, probably mediated by protein factors (Nozaki and Cross, 1995). The 3' untranslated regions (3’UTR) of mRNA are involved in the quantitative and temporal regulation of expression of most stage-specific genes described in kinetoplastids (Vanhamme and Pays, 1995). In most cases, the 3’UTR is involved in controlling the stability of the mRNA, but there is evidence that elements within the 3’UTR interfere with the regulation of several stage-specific genes by modulating the rate of mRNA translation (Berberof et al., 1995; Blattner and Clayton, 1995; Hotz et al., 1997; Boucher et al., 2002). The mechanisms responsible for mRNA translational modulation in kinetoplastids are still not clear. We plan to analyze the mRNA sequences of genes that have their expression controlled at the translational level, and we will search for specific features involved in gene expression modulation. Recently, microarray technology has arisen as a powerful tool to study simultaneously the expression pattern of thousands of genes. We have been using self-made T. cruzi microarrays to determine if polysomal mobilization is an important mechanism for modulating gene expression during metacyclogenesis. Gene expression analysis by competitive hybridization will help us identify and characterize other stage-specific genes that participate in the metacyclogenesis process. Moreover, it will help us to understand the mechanisms responsible for gene expression regulation during T. cruzi metacyclogenesis. ACKNOWLEDGMENTS This work received support from PADCT, PRONEX, CNPq, Fundação Araucária and Fiocruz. A.R. Ávila, V. Monteiro-Góes, B. Dallagiovanna and S. Goldenberg thank CNPq for their fellowships. REFERENCES Abuin, G., Freitas-Junior, L.H., Colli, W., Alves, M.J. and Schenkman, S. (1999). Expression of trans-sialidase and 85-kDa glycoprotein gene in Trypanosoma cruzi is differentially regulated at the post-transcriptional level by labile protein factors. J. Biol. Chem. 274: 13041-13047. Andrade, A.F.B., Esteves, M.J.G., Angluster, J., Gonzalez-Perdomo, M. and Goldenberg, S. (1991). Changes in cell-surface carbohydrates of Trypanosoma cruzi during metacyclogenesis under chemically defined conditions. J. Gen. Microbiol. 137: 2845-2849. Ávila, A.R., Yamada-Ogatta, S.F., da Silva Monteiro, V., Krieger, M.A., Nakamura, C.V., de Souza, W. and Goldenberg, S. (2001). Cloning and characterization of the metacyclogenin gene, which is specifically expressed during Trypanosoma cruzi metacyclogenesis. Mol. Biochem. Parasitol. 117: 169-177. Berberof, M., Vanhamme, L., Tebabi, P. and Pays, E. (1995). The 3'-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. Embo J. 14: 2925-2934. Blattner, J. and Clayton, C.E. (1995). The 3'-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase-encoding genes mediate developmental regulation. Gene 162: 153-156. Böker, C.A. and Schaub, G.A. (1984). Scanning electron microscopic studies of Trypanosoma cruzi in the rectum of its vector Triatomine infestans. Z. Parasitenk. 70: 459-469. Bonaldo, M.C., Scharfstein, J., Murta, A.C. and Goldenberg, S. (1991). Further characterization of Trypanosoma cruzi GP57/51 cystein proteinase as the major antigen expressed by differentiating epimastigotes. Parasitol. Res. 77: 567-571. Boucher, N., Wu, Y., Dumas, C., Dubé, M., Sereno, D., Breton, M. and Papadopoulou, B. (2002). A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3´-untranslated region element. J. Biol. Chem. 277: 19511-19520. Chagas, C. (1909). Nova tripanosomiase humana. Estudo sobre a morfolojia e o cyclo evolutivo do Schizo-trypanum cruzi n.gen.,n.sp.., ajente etiolojico de nova entidade morbida no homem. Mem. Inst. Oswaldo Cruz 1: 159-218. Contreras, V.T., Morel, C.M. and Goldenberg, S. (1985a). Stage-specific gene expression precedes morphological changes during Trypanosoma cruzi metacyclogenesis. Mol. Biochem. Parasitol. 14: 83-96. Contreras, V.T., Salles, J.M., Thomas, N., Morel, C.M. and Goldenberg, S. (1985). In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 16: 315-327. Cooper, R., Inverso, J.A., Espinosa, M., Nogueira, N. and Cross, G.A.M. (1991). Characterization of a candidate gene for GP72, an insect stage-specific antigen of Trypanosoma cruzi. Mol. Biochem. Parasitol. 49: 45-60. Dallagiovanna, B., Plazanet-Menut, C., Ogatta, S.F.Y., Ávila, A.R., Krieger, M.A. and Goldenberg, S. (2001). Trypanosoma cruzi: A gene family encoding chitin-binding-like proteins is post-transcriptionally regulated during metacyclogenesis. Exp. Parasitol. 99: 7-16. de Souza, W. (1984). Cell biology of Trypanosoma cruzi. Int. Rev. Cytol. 86: 197-283. Di Nóia, J.M., Sanchez, D.O. and Frasch, A.C.C. (1995). The protozoan Trypanosoma cruzi has a family genes resembling the mucin genes of mammalian cells. J. Biol. Chem. 270: 24146-24149. Dingwall, C. and Laskey, R.A. (1991). Nuclear targeting sequences - a consensus? Trends Biochem. Sci. 16: 478-481. Esteves, M.G., Gonzalez-Perdomo, M., Alviano, C.S., Angluster, J. and Goldenberg, S. (1998). Changes in fatty acid composition associated with differentiation of Trypanosoma cruzi. FEMS Microbiol. Lett. 59: 31-34. Figueiredo, R.C.B.Q., Rosa, D.S. and Soares, M.J. (2000). Differentiation of Trypanosoma cruzi epimastigotes: metacyclogenesis and adhesion to substrate are triggered by nutritional stress. J. Parasitol. 86: 1213-1218. Fragoso, S.P., Mattei, D., Hines, J.C., Ray, D. and Goldenberg, S. (1998). Expression and cellular localization of Trypanosoma cruzi type II DNA topoisomerase. Mol. Biochem. Parasitol. 94: 197-204. Fragoso, S.P., Plazanet-Menut, C., Carreira, M.A., Motta, M.C., Dallagiovana, B., Krieger, M.A. and Goldenberg, S. (2003). Cloning and characterization of a gene encoding a putative protein associated with U3 small nuclear ribonucleoprotein in Trypanosoma cruzi. Mol. Biochem. Parasitol. 126: 113-117. Fraidenraich, D., Peña, C., Isola, E.L., Lammel, E.M., Coso, O., Añel, A.D., Pongor, S., Baralle, F., Torres, H.N. and Flawia, M.M. (1993). Stimulation of Trypanosoma cruzi adenylate cyclase by an alpha D-globin fragment from Triatoma hindgut: effect on differentiation of epimastigote to trypomastigote forms. Proc. Natl. Acad. Sci. USA 90: 10140-10144. Gale, M., Carter, J.V. and Parson, M. (1994). Translation control mediates the developmental-regulation of the Trypanosoma brucei Nrk protein kinase. J. Biol. Chem. 18: 31659-31665. Garcia, E.S., Gonzalez, M.S., de Azambuja, P., Baralle, F.E., Fraidenraich, D., Torres, H.N. and Flawia, M.M. (1995). Induction of Trypanosoma cruzi metacyclogenesis in the gut of the hematophagous insect vector, Rhodnius prolixus, by hemoglobin and peptides carrying alpha D-globin sequences. Exp. Parasitol. 81: 255-261. Goldenberg, S., Salles, J.M., Contreras, V.T., Lima Franco, M.P., Katzin, A.M., Colli, W. and Morel, C.M. (1985). Characterization of messenger RNA from epimastigotes and metacyclic trypomastigotes of Trypanosoma cruzi. FEBS Lett. 180: 265-270. Gonzales-Perdomo, M., Romero, P. and Goldenberg, S. (1988). Cyclic AMP and adenylate cyclase activators stimulate Trypanosoma cruzi differentiation. Exp. Parasitol. 66: 205-212. Graham, S.V., Wyner, B. and Barry, J.D. (1998). Activity of a Trypanosome metacyclic variant surface glycoprotein gene promoter is dependent upon life cycle stage and chromosomal context. Mol. Cell. Biol. 18: 1137-1146. Heath, S., Hieny, S. and Sher, A. (1990). A cyclic AMP inducible gene expressed during the development of infective stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 43: 133-142. Hotz, H., Hartmann, C., Huober, K., Hug, M. and Clayton, C. (1997). Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3’-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25: 3017-3025. Kollien, A.H., Schmidt, J. and Schaub, G.A. (1998). Modes of association of Trypanosoma cruzi with the intestinal tract of the vector Triatoma infestans. Acta Trop. 70: 127-141. Krieger, M.A. and Goldenberg, S. (1998). Representation of differential expression (RDE): A new approach to study differential gene expression in trypanosomatids. Parasitol. Today 14: 163-166. Krieger, M.A., Ávila, A.R., Ogatta, S.F.Y., Plazanet-Menut, C. and Goldenberg, S. (1999). Differential gene expression during Trypanosoma cruzi metacyclogenesis. Mem. Inst. Oswaldo Cruz 94 (Suppl. 1): 165-168. Nozaki, T. and Cross, G.A. (1995). Effects of 3’untranslated and intergenic regions on gene expression in Trypanosoma cruzi. Mol. Biochem. Parasitol. 75: 55-67. Pays, E. and Nolan, D.P. (1998). Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 91: 3-36. Schaub, G.A. (1989). T. cruzi: quantitative studies of development of two strains in small intestine and rectum of the vector T. infestans. Exp. Parasitol. 68: 260-273. Schenkman, S., Eichinger, D., Pereira, M.E.A. and Nussenzweig, V. (1994). Structural and functional properties of Trypanosoma trans-sialidas. Ann. Rev. Microbiol. 48: 499-523. Schmidt, J., Kleffmann, T. and Schaub, G.A. (1998). Hydrophobic attachment of Trypanosoma cruzi to a superficial layer of the rectal cuticle in the bug Triatoma infestans. Parasitol. Res. 84: 527-536. Sher, A., Crane, M.S.J. and Kirchoff, L.V. (1983). Incubation in mice provides a signal for the differentiation of Trypanosoma cruzi epimastigotes to trypomastigotes. J. Protozool. 30: 278-283. Teixeira, S.M.R. (1998). Control of gene expression in Trypanosomatidae. Braz. J. Med. Biol. Res. 31: 1503-1516. Teixeira, S.M., Kirchhoff , L.V. and Donelson, J.E. (1995). Post-transcriptional elements regulating expression of mRNAs from the amastin/tuzin gene cluster of Trypanosoma cruzi. J. Biol. Chem. 270: 22586-22594. Tomás, A.M. and Kelly, J.M. (1996). Stage-regulated expression of cruzipain, the major cysteine protease of Trypanosoma cruzi is independent of the level of RNA1. Mol. Biochem. Parasitol. 76: 91-103. Tomás, A.M., Miles, M.A. and Kelly, J.M. (1997). Overexpression of cruzipain, the major cysteine proteinase of Trypanosoma cruzi, is associated with enhanced metacyclogenesis. Eur. J. Biochem. 244: 596-603. Vanhamme, L. and Pays, E. (1995). Control of gene expression in trypanosomes. Microbiol. Rev. 59: 223-240. Wallbanks, K.R., Molyneux, D.H. and Dirie, M.F. (1989). Chitin derivates as novel substrates for Trypanosoma brucei brucei attachment in vitro. Acta Trop. 46: 63-68. Zeledon, R., Bolanos, R. and Rojas, M. (1984). Scanning electron microscopic studies of the final phase of the life cycle of Trypanosoma cruzi in the insect vector. Acta Trop. 41: 39-43. |
|