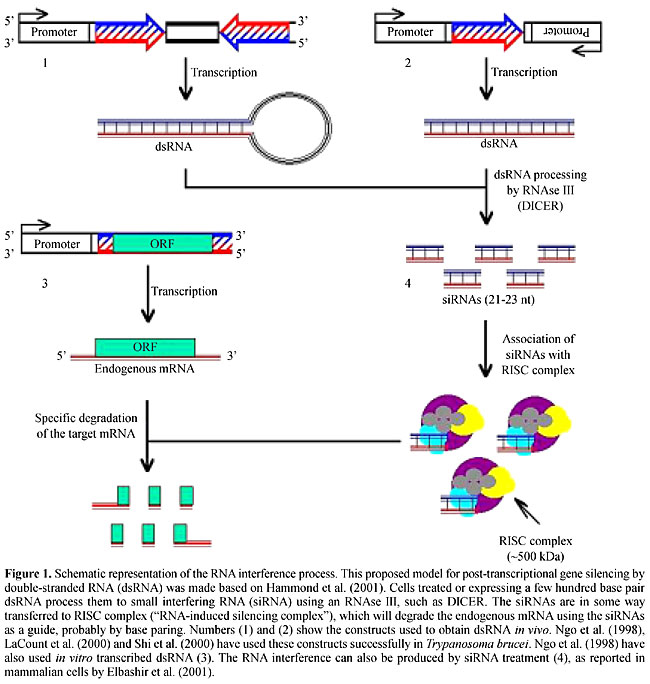
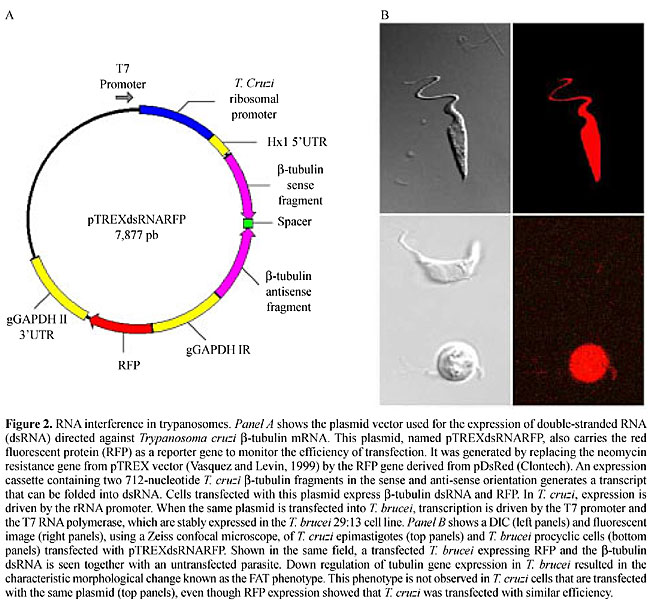
ABSTRACT. Mechanisms controlling gene expression in trypanosomatids depend on several layers of regulation, with most regulatory pathways acting at a post-transcriptional level. Consequently, these parasites can follow the rapid changes associated with transitions between the insect vector and the mammalian host, with instant reprogramming of genetic expression. Using primarily Trypanosoma brucei as a model, the basic controlling mechanisms have been elucidated and now researchers are beginning to define the cellular factors involved in the transcription, processing and translation of the mRNAs in these parasites. We describe some of the studies made on a subset of genes that are differentially expressed during the life cycles of T. brucei and T. cruzi. It is becoming evident that the regulatory strategies chosen by different species of trypanosomatids are not the same, and therefore, the lessons learned from one species do not necessarily apply to the others. Some of the tools available for genetic manipulation that have been developed along with these studies are also described. Two of them are of particular interest in this postgenomic period: inducible systems to express foreign genes and specific inhibition of gene expression by RNA interference. Key words: Trypanosoma, Gene expression, RNA interference, Double-stranded RNA (dsRNA) INTRODUCTION Members of the Trypanosomatidae family constitute a fascinating group of flagellated protozoans; many of them are digenetic parasites whose life cycles present multiple differentiation forms alternating between invertebrate and vertebrate hosts. Trypanosoma brucei, T. cruzi and various species of Leishmania cause a spectrum of parasitic diseases affecting about 550 million people in the developing world (http://www.who.ch). The South American trypanosome, T. cruzi, the etiological agent of Chagas’ disease, multiplies as noninfective epimastigotes in the gut of triatomine bugs and differentiates into nondividing, infective trypomastigotes. In the mammalian host, they invade a variety of cell types and differentiate into a third stage, the intracellular, proliferative amastigotes. Leishmania has an extracellular, elongated form that multiplies in the gut of the insect vector, the sandfly, and a spherical form that is adapted to multiplication in the lysosomes of mammalian macrophages. The African sleeping sickness parasite, T. brucei, is never intracellular, has one form, called procyclic, that multiplies in the gut of the tsetse fly and differentiates into a bloodstream form, which is found in the blood and tissue fluids of mammal hosts. Adaptation to these distinct environments calls for major changes in morphology, surface composition and biochemical pathways, and therefore in gene expression. Being early branching eukaryotes, these microorganisms have attracted the attention of parasitologists, not only for their medical relevance, but also because they present many distinctive features in the mechanisms controlling gene expression. Some of the unusual features found in trypanosomatids include polycistronic transcription, trans-splicing processing of the pre-mRNA, extensive mitochondrial RNA editing, and transcription of protein coding genes by RNA polymerase I. Because primary transcripts are polycistronic, cleavage of the pre-mRNA has to occur in the nucleus in order to produce monocistronic mRNAs. These cleavage reactions are linked to the addition of the 39-nucleotide miniexon (or spliced leader, SL) containing a methylated cap at the 5' end and the poly (A) tail at the 3' end of each mRNA. SL addition results from a transesterification reaction called trans-splicing, which requires a conserved AG dinucleotide as an SL addition site. Studies with various trypanosomatids have provided strong evidence demonstrating that SL addition and polyadenylation are not independent events; instead, they are part of a “cut-and-paste” mechanism that occurs concomitantly or immediately after transcription: poly (A) selection is governed by the location of the SL addition site of the downstream gene in the polycistronic primary transcript. In addition to the correct distance, the presence of a polypyrimidine-rich motif is also crucial, since only AG dinucleotides situated downstream from a polypyrimidine tract are used as SL acceptor sites (LeBowitz et al., 1993; Matthews et al., 1994; for a recent review, see Clayton, 2002). EXPRESSION OF VARIANT SURFACE GLYCOPROTEIN GENES AND THE CONTROL OF ANTIGENIC VARIATION IN TRYPANOSOMA BRUCEI Most of the early studies on gene expression in trypanosomatids were focused on the process of antigenic variation, a powerful survival strategy devised by African trypanosomes, which allows them to escape immunological attack from the host. Variant surface glycoprotein (VSG; Cross, 1975) is a surface molecule known to be the single major protein present in the bloodstream forms of T. brucei. During infection, sequential peaks of parasitemia are due to distinct trypanosome populations, expressing one different VSG on their surface at a time. When host antibodies against one type of VSG molecule are produced and the immune system destroys the trypanosome population expressing this antigen, a few surviving parasites change the type of VSG they express and, with a new surface coat, begin reproducing as if they had never been detected by host-immune defenses. Several interesting questions regarding VSG switching have challenged molecular parasitologists, the first one being: how does a trypanosome switch from the expression of VSG gene A to gene B, while keeping all the other VSG genes silent? The number of VSG genes in the diploid genome of T. brucei has been estimated to be about 1,000. These VSG genes are scattered among one hundred or more minichromosomes (50-150 kb each) and eleven pairs of large megachromosomes (1-6 Mb each) (Borst et al., 1996; Cross, 1996). The work of Williams and collaborators provided the first evidence of rearrangements of VSG genes in the parasite genome (Williams et al., 1979). After the cloning of the first VSG genes, various investigators were able to compare their chromosomal environment and to identify some of the differences among active VSG genes and silent ones. These studies demonstrated that the expressed VSG gene is always located near a chromosome end, about 1-3 kb from the start of a repetitive hexamer present in the telomere, and VSG genes are not alone. Instead, they are part of a long polycistronic operon, called an expression site or ES, that can extend for 50-60 kb and contains about 10 other genes (Cully et al., 1985; Gibbs and Cross, 1988). The products of these genes, called expression site associated genes (ESAG), have no functional relationship to VSG, but for at least two of them, which encode the two subunits of the T. brucei heterodimeric transferrin receptor, this association seems fruitful. The fact that slightly different copies of transferrin receptor are transcribed along with VSG allows the parasite to change the type of receptor that is expressed during an infection (Salmon et al., 1994). Since some VSG genes are also found internally in the chromosomes, some type of DNA rearrangement must occur (such as the ones originally described in Williams et al., 1979) in order to activate those internal genes. But since not all VSGs located in telomers are being expressed in any one cell, there must also exist some type of in situ gene activation/inactivation to differentiate these telomeric ES. So, it appears that a VSG gene has two basic alternatives to be turned on: either it is already in a telomeric ES that needs to be activated in situ (concomitantly with the inactivation of another ES), or it is an internal gene that needs to be copied and moved near to a telomere, where it can replace an active gene (Borst et al., 1996; Cross, 1996). Understanding these mechanisms, particularly the in situ switch, has been a difficult task. The cloning of promoter regions from active VSG genes, crucial for the understanding of the mechanism of VSG regulation, was not an easy task since promoter sequences were found 60 kb upstream of a VSG gene (Zomerdijk et al., 1990; Borst et al., 1996; Cross, 1996). After sequencing several VSG promoters, another unusual discovery was made; they do not possess any of the typical sequences present in eukaryotic RNA pol II promoters, such as TATA box. Moreover, nuclear run-on experiments showed that VSG gene promoters are more related functionally to RNA pol I promoters since they are 100% active in the presence of 1 mg/ml of a-amanitin (Lee and Van der Ploeg, 1997). Thus, it appears that in T. brucei, a unique system is in place, allowing transcription of protein-coding genes to be initiated by RNA polymerase I or an RNA polymerase I-like enzyme. This is only possible in trypanosomes because maturation of the 5’end (and the addition of cap) is independent of the RNA polymerase. Contrary to the general process of eukaryote transcription, in which the enzymes required for the capping process are thought to be associated with RNA polymerase II, in trypanosomes the trans-splicing mechanism provides the SL sequence, which already contains a cap structure at its 5’end. Recently, a major discovery regarding the mechanism of selecting a single VSG for transcription has been reported by Navarro and Gull (2001). Using monoclonal antibodies against the T. brucei RNA pol I, they showed that the active VSG is present in a specific structure in the nucleus of bloodstream trypanosomes, which they called “expression site body”, or ESB. By tagging active VSGs with a green fluorescent protein, they showed that ESB is always associated with an actively expressed copy of the gene. Therefore, because there is only one ESB in each parasite and there is only space for one VSG expression site in each ESB, they explain the selective activation of one VSG at time by a very simple model based on the physical location of the active VSG within this special “transcription factory”. What makes their model even more interesting is that the same principle may be used to explain other examples of monoallelic expression of multigene families, not only in related parasites, such as the case of the var gene family of Plasmodium, but in situations involving more complex genomes, such as the large family of mammalian G-protein coupled olfactory receptor genes (Kratz et al., 2002). POST-TRANSCRIPTIONAL MECHANISMS AS THE MAIN LEVEL OF CONTROL OF GENE EXPRESSION IN TRYPANOSOMES In T. brucei, in addition to VSG, a second group of genes encoding highly abundant surface glycoproteins has been used as a model to explain the mechanisms of gene expression control. When bloodstream trypanosomes differentiate into the procyclic form found in the tsetse fly gut, the VSG coat is rapidly replaced by a new surface coat composed of small, invariant, acidic proteins named EP-procyclin and GPEET-procyclin (for a review, see Roditi et al., 1998), which formerly were called procyclic acid repetitive proteins (PARP). EP-/GPEET-procyclin genes comprise a polymorphic gene family that is found in two genetic loci (a and b) in the genomes of several strains of T. brucei. Two or three EP-/GPEET-procylin genes are found in each locus, together with a EP-/GPEET-procylin-associated gene (PAG). Like the majority of trypanosome genes, EP-/GPEET-procylin genes are part of a polycistronic transcription unit. And like VSG genes, EP-/GPEET-procylin genes are transcribed by an a-amanitin-resistent RNA polymerase (Rudenko et al., 1990). However, sequence comparison shows that the EP-/GPEET-procylin promoters and VSG promoters bear little resemblance to each other, or to the classical RNA polymerase I rRNA promoter (Lee and Van der Ploeg, 1997). Stage-specific control of EP-/GPEET-procylin gene expression occurs at several levels; the increased steady-state levels of procyclin mRNA, seen 2 h after bloodstream forms differentiate into insect forms, is due to a 5- to 10-fold increase in transcription initiation as well as transcription elongation of EP-/GPEET-procylin genes (Roditi, 1996). Although EP-/GPEET-procylin mRNA is not detectable in bloodstream trypanosomes, EP-/GPEET-procylin genes are transcribed at low levels in bloodstream parasites. Rapid degradation, together with inefficient translation in bloodstream forms, accounts for the insect stage-specific expression of EP-/GPEET-procylin genes in T. brucei. Regulatory sequences, such as a 16 mer stem-loop and U-rich 26 mer involved with the differential expression of these genes, have been identified in the 3’UTR of EP-/GPEET-procylin genes. Although EP-/GPEET-procylin and PAG genes are derived from the same polycistronic transcription unit, there is a 100-fold difference in the steady-state levels when EP-/GPEET-procyclin and PAG mRNAs are compared (Roditi et al., 1998). Similar to VSG and ESAG, the expression pattern of the EP-/GPEET-procylin/PAG gene cluster as well as those of other genes in T. cruzi and Leishmania (see below) clearly shows the importance of post-transcriptional mechanisms controlling gene expression. Contrary to VSG and EP-/GPEET-procylin genes of T. brucei, the majority of genes from Trypanosoma and Leishmania are transcribed by an a-amanitin sensible RNA polymerase II. Unexpectedly, although much effort has been expended in the search for an RNA pol II promoter, no sequences displaying typical characteristics of such promoters have been found in trypanosomatids. One of the recent surprises about gene expression in these organisms came from the sequence of chromosome 1 of Leishmania major: the 269-kb chromosome contains 79 genes; 50 of them are lined up one after another in one strand and the other 29 are packed adjacent to each other on the opposite strand (Myler et al., 1999). Although it is believed that the 1.6-kb fragment that separates the two gene clusters has the characteristics of a bi-directional promoter, it is also possible that low levels of transcription initiation occur at nonspecific regions in the genome. Similar types of genomic organization have also been found in other chromosomes of T. cruzi and T. brucei (Andersson et al., 1998; El-Sayed et al., 2000). In contrast to the lack of information about the RNA polymerase II complex in trypanosomes, much attention has been given to the study of regulatory elements derived from untranslated regions of several genes that are submitted to post-transcriptional control. The 3’UTR has emerged as a main site related to mechanisms controlling mRNA stability for several genes (Hehl et al., 1994; Berberof et al., 1995; Furger et al., 1997). Using transient transfections with the CAT or luciferase reporter genes, various researchers have demonstrated the presence of elements in the 3’UTR of several mRNAs, several of them containing U-rich sequences that confer developmental regulation of the reporter genes (Hehl et al., 1994). The two examples below illustrate some of the recent studies in T. cruzi. In this parasites’ genome a tandem array of alternating genes encoding amastin, a surface glycoprotein and tuzin, a G-like protein, are polycistronically transcribed in all three forms of the parasites’ life cycle. In spite of the constitutive transcription, the steady-state levels of amastin genes are 60-fold higher in amastigotes, compared to epimastigote forms. We have shown that the half-life of amastin mRNAs is 7-fold longer in amastigotes than in epimastigotes and that a 630-nucleotide 3’UTR is responsible for amastin up-regulation (Teixeira et al., 1995). This positive effect is likely mediated by a sequence that binds to an RNA stabilizing factor present in amastigotes (Coughlin et al., 2000). Mucin genes are part of an even larger family of cell surface proteins of T. cruzi, with hypervariable regions expressed in various stages of the parasite life cycle. Di Noia et al. (2000) have shown that SMUG mucin mRNA are more abundant in the insect stage and that RNA turnover is controlled by an AU-rich element (ARE) localized in the 3'-UTR. These researchers have also shown that an RNA-binding protein, named TcUBP-1, is involved in mRNA destabilization in vivo through binding to the ARE of SMUG mucin mRNAs. They have gone further in characterizing this trans-acting factor, showing that TcUBP-1 is part of a ~450-kDa ribonucleoprotein complex with a poly (A)-binding protein and a novel 18-kDa RNA-binding protein, named TcUBP-2 (D’Orso and Frasch, 2002). Employing several layers of regulation, these parasites can ensure that rapid changes associated with transmission between insect vector and mammalian host are followed by an instant reprogramming of genetic expression. The examples shown above demonstrate that post-transcriptional control would be a crucial mechanism assuring this regulation. GENETIC MANIPULATION IN TRYPANOSOMES: INTERFERING WITH GENE EXPRESSION Most of the knowledge about the mechanisms of gene expression in trypanosomatids resulted from the development of transfection protocols, which allowed manipulation of genes, generation of knock out mutants and the introduction of genetic markers in the parasite. The first transfection experiments in trypanosomes and Leishmania were performed in the early 90’s in the absence of information regarding the sequences required for gene expression in these organisms (Laban and Wirth, 1989). Soon thereafter, most of the elements involved with gene expression in these parasites were identified, and improved vectors were developed, promoting high levels of expression of foreign genes in various species. These vectors must contain SL addition sites both upstream (for trans-splicing) and downstream (for polyadenylation) from the exogenous gene, as well as promoter sequences. Researchers from several labs have identified sequences derived from various genes that can be used to provide efficient trans-splicing and polyadenylation. On the other hand, we are much more limited in the choice of promoters; while VSG and EP-/GPEET-procyclin have been the favorite sources of strong promoters in T. brucei, for expression vectors in T. cruzi and Leishmania, the only option available is the rRNA promoter. Interestingly, various research groups have shown that it is possible to obtain relatively high levels of expression of foreign genes in T. cruzi and Leishmania using vectors that do not contain promoter sequences (Laban and Wirth, 1989; Teixeira et al., 1995). A major improvement that allowed better control of genetic manipulation in these parasites was the development of inducible expression of gene products with the tetracycline repressor. In this system, which was initially developed for T. brucei, transgenic parasites expressing the tetracycline repressor of E. coli exhibited inducer (tetracycline)-dependent expression of a reporter gene cloned downstream from a trypanosome promoter bearing the tet operator (Wirtz et al., 1999). Following these initial experiments, this prokaryotic inducible system was successfully introduced into T. cruzi and Leishmania donovani (Wen et al., 2001; Yan et al., 2001). In these transfected parasites, reporter gene expression was controlled over a range of four orders of magnitude in response to variations in tetracycline concentration. Now we have a prokaryotic tetracycline-responsive repressor/operator system available for studies in trypanosomes, which has proven to be an excellent tool for dissecting the functions of essential genes and for the expression of toxic gene products. In 1998, Ngo et al., discovered another powerful tool for the genetic manipulation of trypanosomes that allowed the generation of “knock out” mutants by targeting an mRNA through the mechanisms of RNA interference (RNAi). RNAi is a very specific gene silencing mechanism guided by double-stranded RNA (dsRNA) bearing sequences derived from a target gene. This phenomenon was first reported in C. elegans (Fire et al., 1998) and has been described in several other organisms, including T. brucei (reviewed by Ullu et al., 2002). Briefly, exogenously synthesized or internally expressed dsRNA homologous to the coding sequence of a target gene is processed into 20-24-nucleotide long RNAs that work as active guides for mRNA degradation (Figure 1) (Hammond et al., 2000, 2001; Ullu et al., 2002). The exact mechanism of gene silencing by dsRNA is still unknown, and there is much current research towards the characterization of the factors involved. Several reports have described the involvement of a dsRNA nuclease (DICER), a helicase (SMG2), and a protein homologous to an elongation factor (RDE-1) (Tabara et al., 1999; Domeier et al., 2000; Bernstein et al., 2001). Regarding the biological role of RNAi, it has been speculated that naturally occurring dsRNA may be involved in the mechanism for transposon silencing (Ketting et al., 1999; Tabara et al., 1999; Ullu et al., 2002). Researchers have successfully used this powerful technique to “knock out” gene expression in a specific manner in many organisms. As shown in C. elegans, functional genomic analysis by systematic RNA interference is, in principle, an approach that can be applied to any organism that has been brought into the post-genomic era. It is particularly convenient as a methodology to study trypanosomatid genes where antisense RNA has failed and conventional gene knockout is hindered by the fact that most genes are encoded by multiple copy gene families (Fire et al., 1998). Unfortunately, although several reports now show that this approach works well in T. brucei, there have been no reports describing the applicability of RNAi for knocking down genes in Trypanosomatids other than African trypanosomes. Several aspects of T. brucei cell biology and gene function, such as the effect of knocking out a-tubulin (Ngo et al., 1998), the flagellum adhesion glycoprotein-1 (FLA1) (LaCount et al., 2000) and the paraflagellar rod protein (Bastin et al., 2000) have been investigated with RNAi. Ngo et al. (1998) used both in vitro transcribed dsRNAs and plasmid constructs with a- and b-tubulin sequences to generate dsRNAs in T. brucei. Since then, other groups have improved the methodology, using the tetracycline inducible system, to control the production of dsRNA in the parasite, and thus knocking out the target mRNA after the addition of tetracycline (Shi et al., 2000; Wang et al., 2000; LaCount et al., 2002). In spite of the immense success of the RNAi approach for gene studies in T. brucei and, more recently, in the related trypanosome, T. congolense (Inoue et al., 2002), this methodology appears to be slightly more difficult when applied to other members of the trypanosomatid family, such as Leishmania (Zhang and Matlashewski, 2000) and T. cruzi (daRocha, W.D. and Donelson, J.E., unpublished results). Ngo et al. (1998) have shown that knocking out a-tubulin resulted in a characteristic phenotype of T. brucei procyclic cells, named FAT cells. Knocking out the homologue of GP72 in T. brucei, the flagellar adhesion molecule FLA1, results in a parasite with a flagellum detached from the cell body (LaCount et al., 2000). This is the same phenotype observed by Cooper et al. (1993), when the two alleles of the T. cruzi GP72 gene were disrupted by homologous recombination. We have tried to obtain the same phenotypes in T. cruzi, by expressing dsRNA directed against b-tubulin or GP72 (the FLA1 homologue) in transient transfected epimastigotes, with no success; we did not observe any morphological alterations in transfected cells when we used the CL Brener strain (Figure 2). Since we labeled the transfected parasites with the red fluorescent protein gene, cloned in the same vector, we were confident of the efficiency of the transfection. In another set of experiments, we stably expressed dsRNA against amastin mRNA; we also observed that although dsRNA is produced in large amounts in the cells, no substantial reduction in the levels of amastin mRNA is detected (data not shown). Our results are in line with the work described by Zhang and Matlashewski (2000) who showed that dsRNA was unable to modulate the expression of the A2 gene in Leishmania donovani. These results, together with other unpublished reports from various groups indicate that some factor might be missing in the RNAi pathway in T. cruzi and in Leishmania. Continuing efforts are required to elucidate the differences among some of the regulatory pathways present in these organisms. A better understanding of these pathways may provide new insights into the disease they cause and towards the development of new methods of controlling it.
REFERENCES Andersson, B., Aslund, L., Tammi, M., Tran, A.N., Hoheisel, J.D. and Pettersson, U. (1998). Complete sequence of a 93.4-kb contig from chromosome 3 of Trypanosoma cruzi containing a strand-switch region. Genome Res. 8: 809-816. Berberof, M., Vanhamme, L., Tebabi, P., Pays, A., Jefferies, D., Welburn, S. and Pays, E. (1995). The 3'-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosome brucei. EMBO J. 14: 2925-2934. Bastin, P., Ellis, K., Kohl, L. and Gull, K. (2000). Flagellum ontogeny in trypanosomes studied via an inherited and regulated RNA interference system. J. Cell Sci. 113: 3321-3328. Bernstein, E., Caudy, A.A., Hammond, S.M. and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. Borst, P., Rudenko, G., Taylor, M.C., Blundell, P.A., Van Leeuwen, F., Bitter, W., Cross, M. and McCulloch, R. (1996). Antigenic variation in Trypanosomes. Arch. Med. Res. 27: 379-388. Clayton, C.E. (2002). Life without transcriptional control? From fly to man and back again. EMBO J. 21: 1881-1888. Cooper, R., Ribeiro De Jesus, A. and Cross, G.A. (1993). Deletion of an immunodominant Trypanosoma cruzi glycoprotein disrupts flagellum-cell adhesion. J. Cell Biol. 122: 149-156. Coughlin, B.C., Teixeira, S.M., Kirchhoff, L.V. and Donelson, J.E. (2000). Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3'-untranslated region position-dependent cis-element and an untranslated region-binding protein. J. Biol. Chem. 275: 12051-12060. Cross, G.A. (1975). Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71: 393-417. Cross, G.A. (1996). Antigenic variation in trypanosomes: secrets surface slowly. BioEssays 18: 283-291. Cully, D.F., Ip, H.S. and Cross, G.A. (1985). Coordinate transcription of variant surface glycoprotein genes and an expression site associated gene family in Trypanosoma brucei. Cell 42: 173-182. Di Noia, J.M., D’Orso, I., Sanchez, D.O. and Frasch, A.C. (2000). AU-rich elements in the 3'-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 275: 10218-10227. Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L. and Mang, S.E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabdits elegans. Science 289: 1928-1930. D’Orso, I. and Frasch, A.C. (2002). TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 277: 50520-50528. Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. El-Sayed, N.M., Hegde, P., Quackenbush, J., Melville, S.E. and Donelson, J.E. (2000). The African trypanosome genome. Int. J. Parasitol. 30: 329-345. Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. Furger, A., Schurch, N., Kurath, U. and Roditi, I. (1997). Elements in the 3' untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell Biol. 17: 4372-4380. Gibbs, C.P. and Cross, G.A. (1988). Cloning and transcription analysis of a variant surface glycoprotein gene expression site in Trypanosoma brucei. Mol. Biochem. Parasitol. 28: 197-206. Hammond, S.M., Bernstein, E., Beach, D. and Hannon, G.J. (2000). An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404: 293-296. Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. and Hannon, G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. Hehl, A., Vassella, E., Braun, R. and Roditi, I. (1994). A conserved stem-loop structure in the 3' untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 91: 370-374. Inoue, N., Otsu, K., Ferraro, D.M. and Donelson, J.E. (2002). Tetracycline-regulated RNA interference in Trypanosoma congolense. Mol. Biochem. Parasitol. 120: 309-313. Ketting, R.F., Haverkamp, T.H., van Luenen, H.G. and Plasterk, R.H. (1999). Mut-7 C. elegans, required for transposon silencing and RNA interference, is a homolog of Wener syndrome helicase and RNaseD. Cell 99: 133-141. Kratz, E., Dugas, J.C. and Ngai, J. (2002). Odorant receptor gene regulation: implications from genomic organization. Trends Genet. 18: 29-34. Laban, A. and Wirth, D.F. (1989). Transfection of Leishmania enriettii and expression of chloramphenicol acetyltransferase gene. Proc. Natl. Acad. Sci. USA 86: 9119-9123. LaCount, D.J., Bruse, S., Hill, K.L. and Donelson, J.E. (2000). Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111: 67-76. LaCount, D.J., Barrett, B. and Donelson, J.E. (2002). Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277: 17580-17588. LeBowitz, J.H., Smith, H.Q., Rusche, L. and Beverley, S.M. (1993). Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7: 996-1007. Lee, M.G. and Van der Ploeg, L.H. (1997). Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Ann. Rev. Microbiol. 51: 463-489. Matthews, K.R., Tschudi, C. and Ullu, E. (1994). A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8: 491-501. Myler, P.J., Audleman, L., deVos, T., Hixson, G., Kiser, P., Lemley, C., Magness, C., Rickel, E., Sisk, E., Sunkin, S., Swartzell, S., Westlake, T., Bastien, P., Fu, G., Ivens, A. and Stuart, K. (1999). Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96: 2902-2906. Navarro, M. and Gull, K. (2001). A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414: 759-763. Ngo, H., Tschudi, C., Gull, K. and Ullu, E. (1998). Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95: 14687-14692. Roditi, I., Furger, A., Ruepp, S., Schurch, N. and Butikofer, P. (1998). Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 91: 117-130. Rudenko, G., Le Blancq, S., Smith, J., Lee, M.G., Rattray, A. and Van der Ploeg, L.H. (1990). Procyclic acidic repetitive protein (EP-/GPEET-PROCYCLIN) genes located in an unusually small alpha-amanitin-resistant transcription unit: PARP promoter activity assayed by transient DNA transfection of Trypanosoma brucei. Mol. Cell. Biol. 10: 3492-3504. Salmon, D., Geuskens, M., Hanocq, F., Hanocq-Quertier, J., Nolan, D., Ruben, L. and Pays, E. (1994). A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78: 75-86. Shi, H., Djikeng, A., Mark, T., Wirtz, E., Tschudi, C. and Ullu, E. (2000). Genetic interference in Trypanosoma brucei by heritable inducible double-stranded RNA. RNA 6: 1069-1076. Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123-132. Teixeira, S.M., Kirchhoff, L.V. and Donelson, J.E. (1995). Post-transcriptional elements regulating expression of mRNAs from amastin/tuzin gene cluster of Trypanosoma cruzi. J. Biol. Chem. 270: 22586-22594. Ullu, E., Djikeng, A., Shi, H. and Tschudi, C. (2002). RNA interference: advances and questions. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357: 65-70. Vasquez, M.P. and Levin, M.J. (1999). Functional analysis of the intergenic regions of TcP2b gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene 239: 217-225. Wang, Z., Morris, J.C., Drew, M.E. and Englund, P.T. (2000). Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. Proc. Natl. Acad. Sci. USA 275: 40174-40179. Wen, L., Xu, P., Benegal, G., Carvalho, M.R.C., Butler, D.R. and Buck, G.A. (2001). Trypanosoma cruzi: Exogenously regulated gene expression. Exp. Parasitol 97: 196-204. Williams, R.O., Young, J.R. and Majiwa, P.A. (1979). Genomic rearrangements correlated with antigenic variation in Trypanosoma brucei. Nature 282: 847-849. Wirtz, E., Leal, S., Ochatt, C. and Cross, G.A.M. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99: 89-101. Yan, S., Myler, P.J. and Stuart, K. (2001). Tetracycline regulated gene expression in Leishmania donovani. Mol. Biochem. Parasitol. 112: 61-69. Zhang, W.W. and Matlashewski, G. (2000). Analysis of antisense and double stranded RNA downregulation of A2 protein expression in Leishmania donovani. Mol. Biochem. Parasitol. 107: 315-319. Zomerdijk, J.C., Ouellette, M., ten Asbroek, A.L., Kieft, R., Bommer, A.M., Clayton, C.E. and Borst, P. (1990). The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 9: 2791-2801. |
|