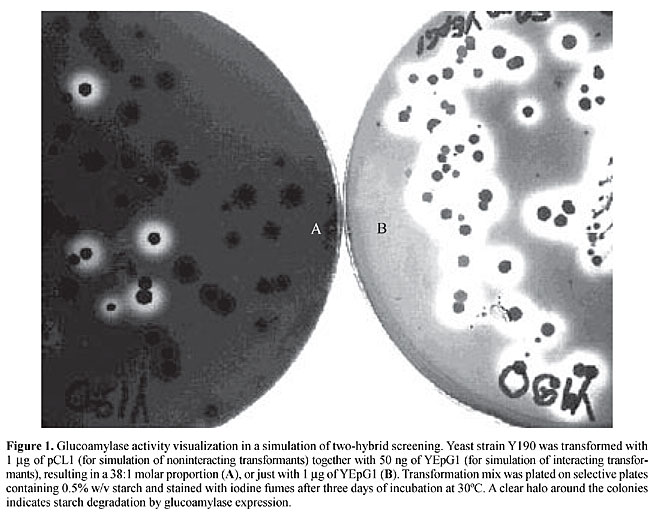
ABSTRACT. The yeast two-hybrid system is a powerful tool for screening protein-protein interactions and has also been used for large-scale studies. We evaluated two protein-coding sequences as reporter genes for the yeast two-hybrid system, to determine if it was suitable as an alternative screening strategy. Aspergillus awamori glucoamylase activity results in clear haloes around colonies producing this enzyme after growth on starch plates and staining with iodine vapors. However, transcription activation by Gal4 on Gal-regulated promoters was insufficient for this type of phenotypic visualization. A modified gene of Aequoria victoria enhanced green fluorescent protein (EGFP) was tested to determine its suitability for interaction screenings with flow cytometry. When the EGFP reporter gene system was incorporated into the cells, Gal4 transcriptional activation produced sufficient fluorescence for detection with the flow cytometer, especially when there were strong interactions. Key words: Yeast two-hybrid system, Reporter genes, Glucoamylase, Green fluorescent protein INTRODUCTION Protein interactions are fundamental to essential processes, such as transcription, translation, signal transduction and cell-cycle control. The yeast two-hybrid system has been used since 1989 (Fields and Song, 1989) to screen for proteic interaction partners in vivo. However, several types of false-positive results have been described in this system (Gietz et al., 1997; Colas and Brent, 1998; Serebriiskii et al., 2000). Despite significant amounts of false positives using two-hybrid screenings, this system has been used for comprehensive analysis of protein-protein interactions since the beginning of 2000 (Uetz et al., 2000). The use of different reporter genes with distinct TATA box and surrounding sequences would avoid the selection of clones whose activation is due to prey interaction with DNA upstream of the reporter gene (Colas and Brent, 1998) instead of bait-prey interaction. Also, false-positive results, due to an eventual increase of basal reporter gene transcription over the phenotypic threshold (without any other support for bait-prey interaction; Colas and Brent, 1998), can be discriminated using reporter genes with different levels of sensitivity. Most two-hybrid host yeast strains have two reporter genes: one auxotrophic marker for screening on selective agar plates and lacZ for qualitative and quantitative evaluation of activation. Even with two reporter genes, the two phenotypes are not scored at the same time, because when the auxotrophic marker is used to screen interacting proteins from a library it is very expensive to check simultaneously for b-galactosidase activity. Moreover, Serebriiskii et al. (2000) described some false positives that induce altered growth and cell permeability in yeast, causing a bias in the perceived activity of LacZ reporters. For these reasons, it would be useful to have a system in which both reporter phenotypes would be visualized at the same time, at low cost and without any bias provoked by alterations in permeability. Amylolytic enzymes are good candidates for reporter phenotypes. They are exported enzymes, their substrate is inexpensive and substrate degradation is easily determined by exposure to iodine. Aspergillus awamori glucoamylase (a1,4-D-glucan-glucohydrolase) is an exohydrolase that catalyses glucose residue liberation from nonreducing ends of starch and other related polymers (Yamasaki et al., 1977). It has been successfully used as a reporter for promoter screening (Scorpione et al., 1993) and for this reason it was evaluated as a two-hybrid reporter gene. Green fluorescent protein (GFP), a 238-amino acid polypeptide, is intrinsically fluorescent, thus, it does not need substrates or co-factors to produce a green emission when excited with UV light (Chalfie et al., 1994). GFP has been used as reporter gene in various organisms, including Escherichia coli, Caernorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, Aedes aegypti, fish, viruses, mammals and plants (Chalfie et al., 1994; Cubbit et al., 1995; Amsterdam et al., 1995; Baulcombe et al., 1995; Plautz et al., 1996; Higgs et al., 1996; Chiu et al., 1996). A GFP-modified version, with the alterations Ser 65 to Thr and Phe 64 to Leu, was named EGFP (enhanced green fluorescent protein; Cormack et al., 1996). EGFP produces fluorescence 35 times more intense than the wild type and has better solubility, as well as faster folding and chromophore maturation (Kain and Ma, 1999). A fluorescent two-hybrid reporter gene might allow screening with a flow cytometer; we tested this system to determine if this screening method would work. MATERIAL AND METHODS Reporter constructions The strategies to build the reporter genes are described in the results section. Molecular biology procedures were implemented as described by Sambrook et al. (1989) and the basic methodology for the experiments with yeast was as described by Guthrie and Fink (1991). PCR amplification of the CYC1 promoter was done with the following primers: 5’ GAC TCC GAG CTC GCA CTC GAG ATC GAC TAT ATA TAT GTC TG 3’ and 5’ CGC GGC ACT AGT ATT AAT TTA GTG TGT GT 3’, and 25 cycles of 1 min at 94ºC for denaturing, 1 min for annealing at 55ºC and 2 min of extension at 72ºC. The upper strand sequence of the GALUAS oligonucleotide is 5’ CGG AGT ACT GTC CTC CGC TCG AGC GGA GTA CTG TCC TCC GAA CGG AGT ACT GTC CTC CGG 3’, and the lower strand sequence is 5’ GAT CCC GGA GGA CAG TAC TCC GTT CGG AGG ACA GTA CTC CGC TCG AGC GGA GGA CAG TAC TCC GAG CT 3’. The lower strand was phosphorylated before annealing with the upper strand. After annealing they were incubated with T4 ligase for dimerization and then phosphorylated to allow ligation with the plasmid. Sequencing Sequencing reactions were conducted using M13 universal primers (Pharmacia Biotech) and the Thermosequence fluorescent-labeled primer cycle sequencing kit (Amersham-Life Science). Sequencing gels were read in an Automated Fluorescent Laser from Pharmacia Biotech. Glucoamylase phenotype Cells were plated on 0.1 up to 2% starch selective media and incubated at 30ºC for 36 h. One half gram of iodine crystals was put on the lids of inverted plates and the plates incubated for up to 3 min, time enough to stain the remaining starch on the plates and to reveal the degradation haloes. Fluorescence analysis Flow cytometry was performed using an FACSvantage cell sorter from Becton and Dickinson Immunocytometry Systems (San Jose, CA, USA). Samples were diluted in 1X PBS to a concentration of 106 cells/ml. The cells were passed at 105 cell/s; fluorescence was detected in FL1 after excitation at 488 nm. For cell sorting, the 35% most fluorescent events were gated using a flow rate of 3,500-4,000 cells/s, and NORMAL-R sort mode. Analysis was performed using the Cell Quest program from BD Sciences. RESULTS Glucoamylase as a two-hybrid reporter gene Formation of translucid haloes on a starch plate stained with iodine when cells are expressing glucoamylase could be a useful phenotype for an additional reporter for two-hybrid screenings. However, it is necessary to make sure that the haloes are readily detectable on transformation selective plates also containing nonhalo-producing clones. To test this system, a two-hybrid host yeast strain (Y190, Clontech) was transformed with both YEpG1 (Scorpione et al., 1993), which constitutively expresses glucoamylase, and pCL1 (Clontech); the latter clone has the same genetic marker but encodes full length Gal4, therefore without any amylolytic activity at a molar proportion of 1:38. There was a clear distinction between glucoamylase-expressing and nonexpressing colonies (Figure 1). There was complete viability of the selected clones after staining (results not shown).
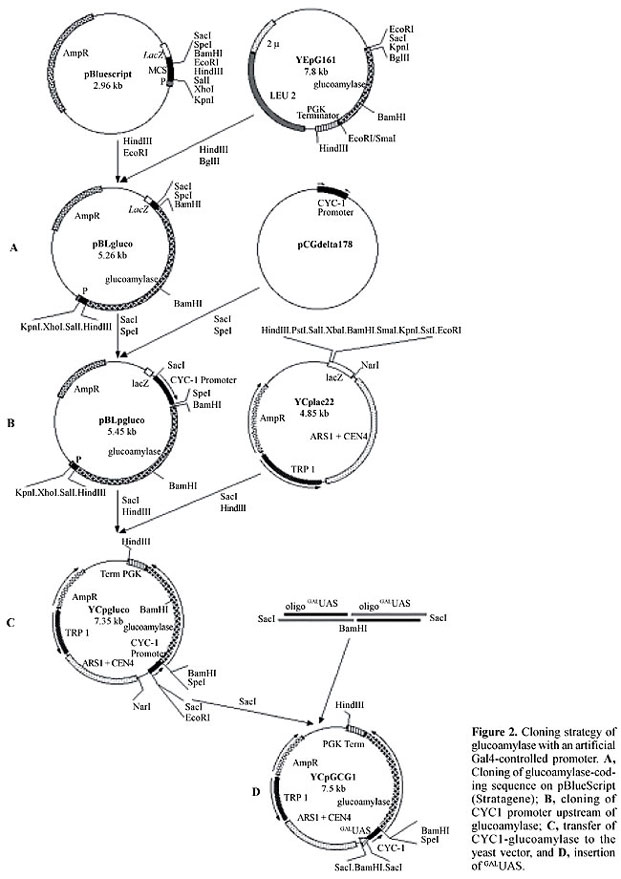
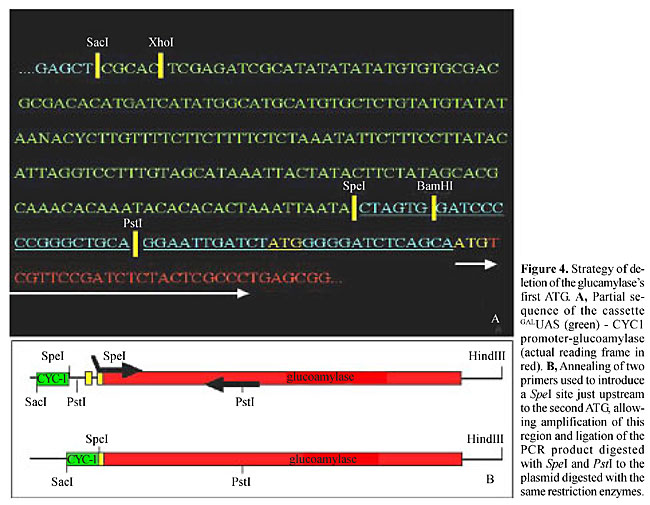
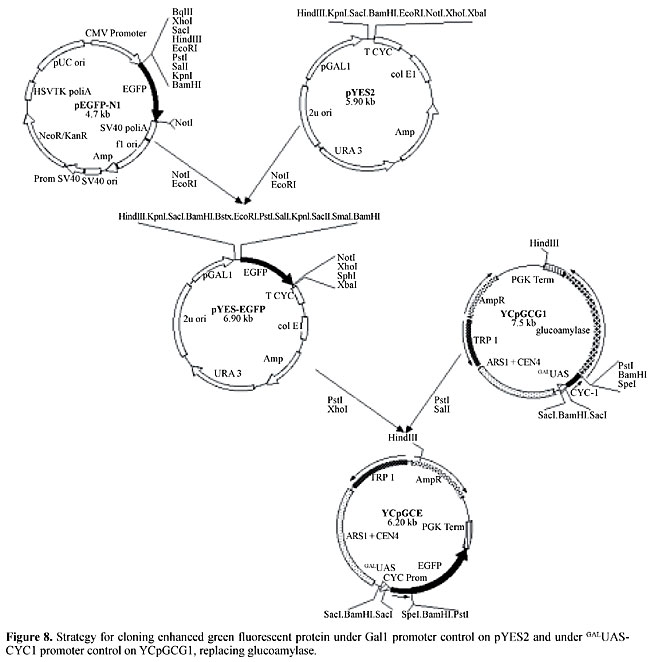
Reporter construction was done as represented in Figure 2, resulting in a centromeric plasmid with a cassette of 6 GALUAS sequences, a CYC1 promoter with a transcriptional start site and a glucoamylase-coding sequence (YCpGCG1). This plasmid was introduced into Y190 cells together with pCL1 (expressing full-length activator Gal4). Even though this assay was designed for maximum activation of reporter transcription, no halo was observed in the iodine-dyed starch plates in which the double transformants were cultivated. Partial plasmid sequencing showed that there was no failure in the junctions in the constructed cassette; however, there was an additional start codon (ATG), outside of the reading-frame, before the functional glucoamylase ATG (Figure 3).
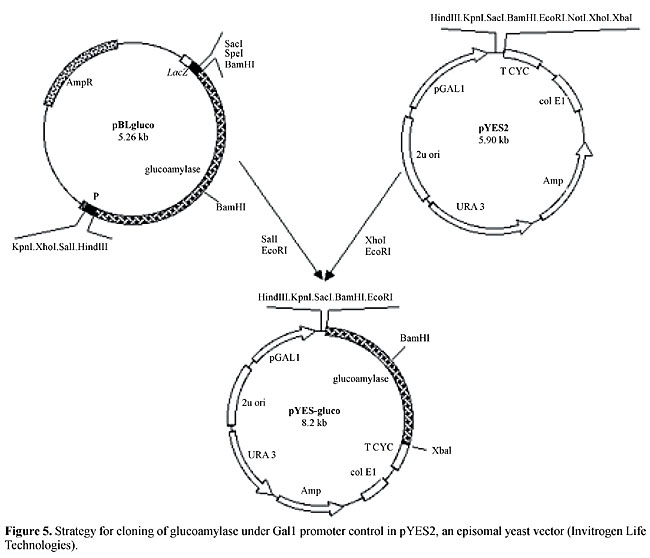
This additional ATG was removed using the strategy depicted in Figure 4. Nevertheless, Gal4 activation did not result in halo visualization. The glucoamylase-coding sequence was then cloned under the control of the Gal1 promoter in the episomal vector pYES2 from Invitrogen Life Technologies (Figure 5). The resulting plasmid, pYES-gluco, still did not confer an amylolytic phenotype to YJ0Z cells (Leuther and Johnston,1992) under Gal4 activation. Therefore, even with multiple copies in an episomal plasmid, the glucoamylase gene was not able to produce a detectable phenotype in a typical two-hybrid assay (e.g., a GALUAS-driven promoter), possibly because it requires a very high transcription level to sufficiently digest starch to create a visible halo. In conclusion, glucoamylase is not a suitable reporter gene for two-hybrid screenings.
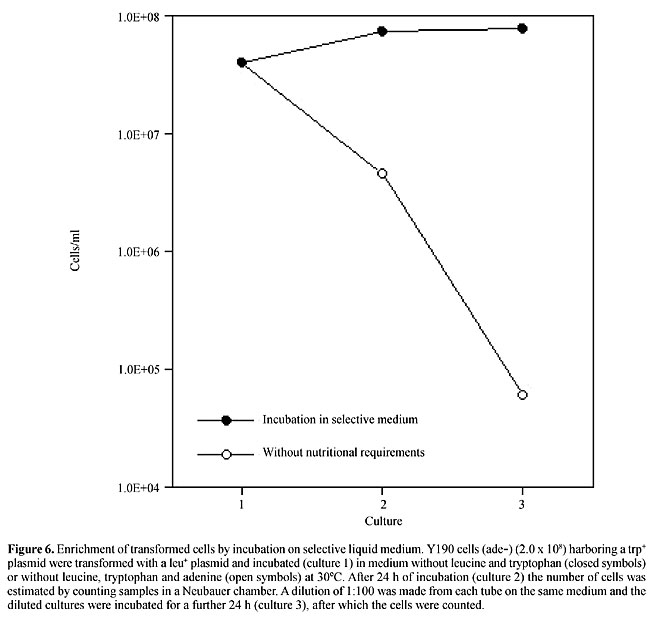
Green fluorescent protein as a two-hybrid reporter gene Fluorescent proteins, especially those that do not require exogenous substrates, can be very useful for expression screenings when a flow cytometer is available. But the screening procedure by the cell sorter should not be done immediately after transformation because the flow cytometer will have to analyze all the cells, including the nontransformed ones, which make up the majority. Therefore, before trying GFP in a two-hybrid simulation, we developed a protocol for enrichment of transformant cells. This protocol is attractive and potentially faster than selection on plates. It was observed that incubation at 30ºC in selective liquid medium for 24 h of 108 cells inoculated just after transformation gave 16 times more cells under microscopy than the culture incubated without the nutritional requirements (a nongrowing culture). If a 1:100 dilution is made after the first 24 h and the tubes are incubated for a further 24 h with the same medium, the difference rises to 1,200 times (Figure 6). With this enrichment of transformed cells, it is possible to conduct screenings using a flow cytometer.
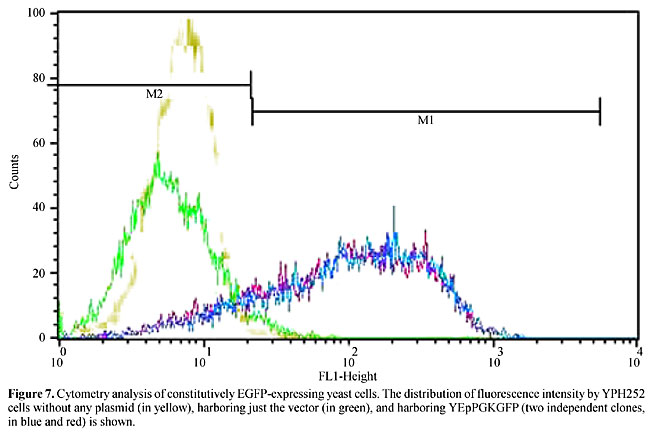
On the other hand, the ability to detect GFP fluorescent yeast cells was checked by expressing EGFP (Cormack et al., 1996), isolated as a BglII/NotI fragment from pEGFP-N1 (Clontech), with the strong yeast promoter PGK (phosphoglycerokinase), isolated as a BamHI/BglII fragment from YEpG1 (Scorpione et al., 1993). These fragments were introduced into YEp161 (Scorpione et al., 1993) after deletion of a 2.1-kb BglII/SmaI fragment encoding for glucoamylase, originating the plasmid YEpPGKGFP. FACS analysis of two yeast clones harboring YEpPGKGFP showed that most of those cells had more intense fluorescence than the control (Figure 7), which would allow them to be sorted. To test this, these cells (which are Ade+) were mixed with another yeast strain (YJ0Z/pCL1, ade2), which does not express EGFP, at a proportion of 30 EGFP-expressing cells to 106 nonexpressing cells, followed by a cell sorting procedure using FACSvantage. Fluorescent cells were collected and incubated in selective media for maintenance of the plasmid for two days as standardized, when around 1,000 cells were plated. After 72 h of incubation it was possible to distinguish ade2 (pink, 17%) from Ade+ (white, 83%) colonies, showing the enrichment of fluorescent cells by just one round of sorting. This proportion was confirmed by analysis of b-galactosidase activity in the colonies (data not shown), which is only presented by YJ0Z/pCL1.
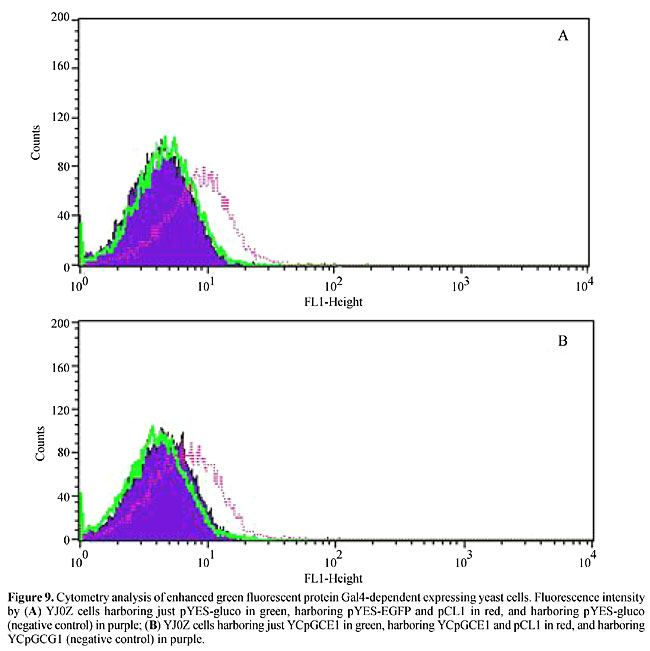
EGFP-coding sequence was cloned into pYES and into YCpGCG1 in substitution of the glucoamylase sequence (Figure 8). Both constructs, when transformed into YJ0Z/pCL1 cells, which express full-length Gal4, produced fluorescent cells at higher rates than the control (Figure 9), allowing them to be sorted from the nonfluorescent cells, but probably with much less enrichment than in the procedure described above, since the expression levels are considerably higher under the control of a strong promoter. We believe that it is possible to use EGFP as a reporter for two-hybrid screenings, especially when a strong interaction is sought. The experimental procedure of enriching transformed cells that is shown here is feasible, opening the perspective of substituting plating for screening assays with rapid cell sorting of interacting clones.
DISCUSSION AND CONCLUSION Glucoamylase and EGFP were tested as reporter genes for a yeast two-hybrid system. Although amylolytic enzymes are potential candidates for reporter phenotypes, Aspergillus awamori glucoamylase did not express a conspicuous phenotype in a two-hybrid reporter construction. The possibility that an error or improper technique during the construction of the expression cassette made it nonfunctional can be discarded, because the same controlling cassette was successfully used to express EGFP under Gal4 control. It seems that glucoamylase’s amylolytic activity is not strong enough to generate clear haloes, unless expressed under very strong promoter control, such as PGK. One reason for this could be the high level of activity needed to significantly reduce the size of starch molecules to prevent iodine staining, since glucoamylase is an exohydrolase. On the other hand, glucoamylase appears to be a good reporter for screening strong promoters. EGFP presented good potential as a two-hybrid reporter gene, although flow cytometry works best for screening for strong interactions. Changing promoters, increasing the number of UAS copies, or using improved versions of fluorescent green protein may increase the sensitivity of this reporter. ACKNOWLEDGMENTS Research supported by FAPEMIG (grant No. CBS 1200/95). A.L. Starling was the recipient of a CAPES fellowship. REFERENCES Amsterdam, A., Lin, S. and Hopkins, N. (1995). The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Devel. Biol. 171: 123-129. Baulcombe, D.C., Chapman, S. and Santa Cruz, S. (1995). Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7: 1045-1053. Chalfie, M., Yu, T., Guskirchen, G., Ward, W.W. and Parsher, D.C. (1994). Green fluorescent protein as a marker for gene expression. Science 263: 802-805. Chiu, W.-L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. and Sheen, J. (1996). Engineered GFP as vital reporter in plants. Curr. Biol. 6: 325-330. Colas, P. and Brent, R. (1998). The impact of two-hybrid and related methods on biotechnology. Trends Biotechnol. 16: 355-363. Cormack, B.P., Valdivia, R.H. and Falkow, S. (1996). FACS-optimized mutants of green fluorescent protein (GFP). Gene 173: 33-38. Cubbit, A.B., Heim, R., Adams, S.R., Boyd, A.E., Gross, L.A. and Tsien, R.Y. (1995). Understanding improving and using green fluorescent proteins. Trends Biochem. Sci. 20: 448-455. Fields, S. and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340: 245-246. Gietz, R.D., Triggs-Raine, B., Robbins, A., Graham, K.C. and Woods, R.A. (1997). Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol. Cell. Biochem. 172: 67-79. Guthrie, C. and Fink, G.R. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol. 194: 3-21, 182-187. Higgs, S., Traul, D., Davis, B.S., Kamrud, K.I., Wilcox, C.L. and Beaty, B.J. (1996). Green fluorescent protein expressed in living mosquitoes - without the requirement of transformation. BioTechniques 21: 660-664. Kain, S.R. and Ma, J.-T. (1999). Green fluorescent protein. Methods Enzymol. 302: 38-43. Leuther, K.K. and Johnston, S.A. (1992). Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science 256: 1333-1335. Plautz, J.D., Day, R.N., Dailey, G.M., Welsh, S.B., Hall, J.C., Halpain, S. and Kay, S.A. (1996). Green fluorescent protein and its derivatives as versatile markers for gene expression in living Drosophila melanogaster, plant and mammalian cells. Gene 173: 83-87. Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989). Molecular Cloning: A laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. Scorpione, R.C., Camargo, S.S., Schenberg, A.C.G. and Astolfi-Filho, S. (1993). A new promoter probe vector for Saccharomyces cerevisiae using fungal glucoamylase cDNA as the reporter gene. Yeast 9: 599-605. Serebriiskii, I., Estojak, J., Berman, M. and Golemis, E.A. (2000). Approaches to detecting false positives in yeast two-hybrid systems. BioTechniques 28: 328-336. Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S. and Rothgerg, J. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. Yamasaki, Y., Suzuki, Y. and Osawa, J. (1977). Three forms of a-glucosidade and glucoamylase from Aspergillus awamori. Agric. Biol. Chem. 41: 2149-2161. |
|