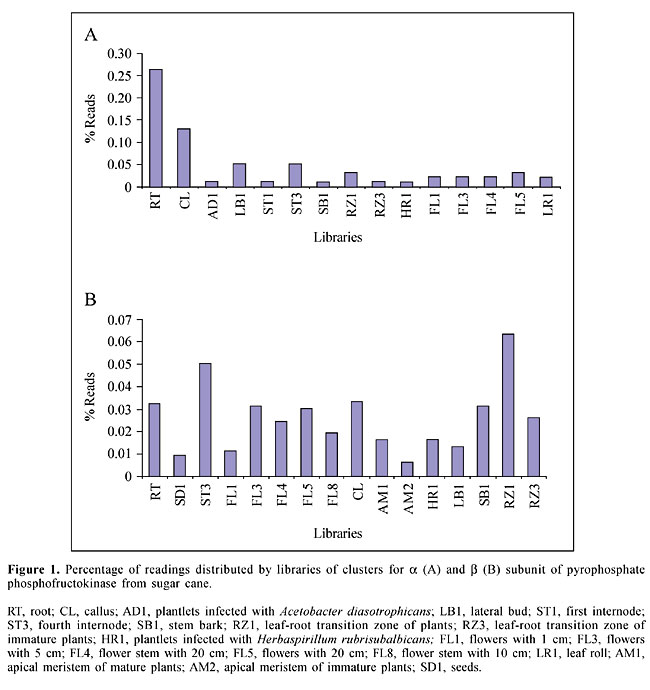
ABSTRACT. Pyrophosphate-dependent phosphofructokinase (PPi-PFK) has been detected in several types of plant cells, but the gene has not been reported in sugar cane. Using Citrus paradisi PPi-PFK gene (AF095520 and AF095521) sequences to search the sugar cane EST database, we have identified both the a and b subunits of this enzyme. The deduced amino acid sequences showed 76 and 80% similarity with the corresponding a and b subunits of C. paradisi. A high degree of similarity was also observed among the PFK b subunits when the alignment of the sugar cane sequences was compared to those of Ricinus communis and Solanum tuberosum. It appears that a and b are two distinct subunits; they were found at different concentrations in several sugar cane tissues. It remains to be determined if the different gene expression levels have some physiological importance and how they affect sucrose synthesis, export, and storage in vacuoles. A comparison between the amino acid sequences of b PFKs from a variety of organisms allowed us to identify the two critical Asp residues typical of this enzyme’s activity site and the other binding sites; these residues are tightly conserved in all members of this protein family. Apparently, there are catalytic residues on the b subunit of the pyrophosphate-dependent enzyme. Key words: Pyrophosphate phosphofructose 1-kinase, Sugar cane, ATP, Sucrose, Sucrose synthase, PPi-PFK INTRODUCTION The timing of the highest levels of sucrose storage in mature sugar cane internodes is of utmost importance for yield performance; it is also a signal for the harvest time. However, sucrose storage in vacuoles is preceded by an intense requirement for metabolic energy, which is mainly provided by sucrose coming from the green leaves. Recently, in addition to the well-known essential role of soluble sugars in metabolism, they have also been recognized as signal molecules (Sheen et al., 1999), regulating sugar-responsive gene expression (Koch, 1996; Smeekens, 1998; Gibson and Grahan, 1999; Gibson, 2000). Absorbing sunlight energy, producing sugars, and sensing a variety of chemical signals (Braam et al, 1997), the higher plants perceive and react accurately to all external environmental stimuli, leading to rapid adjustment of growth according to environmental conditions (Ikeda et al., 1999; Yu, 1999). There is an interrelationship between morphogenesis, environment, and sugar metabolism (Sattler and Rutishauser, 1997). The great variety of biochemical reactions that comprise the synthesis of sucrose and its assimilation into the plant tissues essentially depends on the activities of the enzymes: fructose-1,6-biphosphatase (E.C.3.1.3.11) and sucrose phosphate synthase (E.C.2.4.1.14). But they are not the only relevant enzymes in this process. The enzyme, pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PPi-PFP) (E.C.2.7.1.90), previously described in sugar-cane tissues (Lingle and Smith, 1991), seems to play a crucial role at the regulatory site in plant carbohydrate metabolism (Heldt, 1997). This might be a key part of the process by which plants adjust their growth as a function of sucrose synthesis, export, import, and utilization. Since PPi-PFP is able to accept PPi as a phosphoryl donor, apparently cytosolic pyrophosphate (the supposed cellular by-products), normally found at high concentrations (up to 0.3 mM) in plant cells (Stit, 1990), can mimic the ATP molecule. In addition, it has also been shown that the reaction catalyzed by this enzyme is thermodynamically fully reversible, allosterically modulated (Nielsen, 1995), and should be able to replace both ATP-PFK and FBPase in their typical counter-reaction. Thus, based on strong evidence, it is thought that PPi-PFP is able to provide additional economy of ATP in glycolysis reactions (Plaxton, 1996), and as it modulates the mass diversion of sugar phosphates in plant growth metabolism, this enzyme has an important effect on sugar cane yield. Although PPi-PFP has been identified in a number of plants (Carlisle et al., 1990; Cheng and Tao, 1990; Nielsen, 1995; Theodorou and Plaxton, 1996; Farré et al., 2000) and in microorganisms (Nadkarni et al., 1984; Heinisch et al., 1989; Deng et al., 1998; Rodicio et al., 2000; Chi et al., 2001), its physiological role is still unclear. Early studies have revealed a wide variation in PPi-PFP protein motifs, which vary from small ones, consisting of dimeric forms with a 60-kDa subunit, found in wheat seedlings (Yan and Tao, 1984), to other forms of complex tetrameric proteins consisting of two a and two b subunits that have also been identified in wheat seedlings (Yan and Tao, 1984), potato tubers (Kruger and Denis, 1987), barley seedlings (Nielsen, 1995), and heterooctameric forms composed of four a and four b subunits in yeast cells (Heinisch et al., 1989; Rodicio et al., 2000), and Brassica nigra cells (Theodorou and Plaxton, 1996). The PPi-PFP gene has been cloned and sequenced (Heinisch et al., 1989), providing a basis to identify this gene and to compare the amino acid sequences of both subunits of this enzyme. We examined the cDNA sequence of PPi-dependent phosphofructokinase in a sugar cane EST database, based on gene homology with other crops. MATERIAL AND METHODS Nucleotide sequence of the pfp gene Identification of PPi-dependent phosphofructokinase (PPi-PFK or PFP, E.C.2.7.1.90) a (regulatory) and b (catalytic) gene subunits was made using a database generated by sequencing several different cDNA libraries from the Sugarcane Genome Project (SUCEST), using the amino acid sequence from PPi-dependent phosphofructokinase in Citrus paradisi (GenBank accession numbers AF095520 and AF095521). Processing and annotation Database search for a similar nucleotide sequence was performed by running the tBLASTX program (Altschul et al., 1997, without filter) on the nonredundant nucleotide sequence database from the National Center for Biotechnology Information (NCBI), GenBank, compared with the Sugarcane EST Program (SUCEST) database. The resulting top match clusters with e-values 1-5 or better and 70% identity or better were selected. Following submission, the hits with good quality and similarity were used as queries for another search back to the NCBI database, in order to determine clone identity and to find possible matches. RESULTS Although over 15 clusters each were detected when PPi-dependent phosphofructokinase a and b subunits were used for the search, only about 30% gave e-values over 1-5. By analyzing these, it was found that some clusters were highly similar to the N-terminal portion of the sequences, and are probably full-length cDNAs, although their complete nucleotide sequences are not available at this time. Two clusters similar to the N-terminal portion of the gene pyrophosphate-dependent phosphofructokinase for the a and b PFP-subunits considered to be catalytic (Wang and Shi, 1999) were chosen from the EST database. These were SCCCCL3001B07 and SCCCCL4011H08, respectively. Both clusters cover over 77% of the message, which gives a good degree of confidence for gene identity. These clusters were composed of the alignment of several readings from different clones isolated from various libraries. A wide distribution of these readings among different libraries was observed for both clusters (Figure 1A and B). Clones for the a PFK subunit were detected more frequently in roots and calluses, although they also appeared at a lower frequency in other libraries (Figure 1A). For instance, the clones that express the b subunit were detected at a higher frequency in most of the libraries, when compared to the a subunit from stem tissue clones for this specific gene, although they appeared in roots, flowers, calluses, stem bark, and roots at similar frequencies (Figure 1B).
The predicted amino acid sequences of the PFK b subunit of the EST cluster was compared to the predicted amino acid sequence of R. communis (Genbank: Q41141), Citrus paradisi (Genbank: AF095520) and Solanum tuberosum (Genbank: P21343) (Figure 2). The comparison reveals the existence of active and ligand sites that are highly conserved in all members of the PPi-PFK protein family.
DISCUSSION PPi-PFK is widespread in almost all organisms, from bacteria to higher eukaryotes, and its genomic sequences have been cloned and sequenced, providing a basis for sequence comparisons between different organisms. The amino acid comparison between the clusters found in the search with pyrophosphatase-dependent phosphofructokinase a and b subunits revealed open reading frames (ORFs) with similarities in the N-terminal portion of the protein region, showing that we were able to detect full-length cDNA of this enzyme’s a and b subunits. The categorization group designed from the SUCEST program has already categorized these clusters as part of C-compound and carbohydrate metabolism. Even though the PPi-PFK amino acid sequence is not 100% identical, the residues involved in the active site and the ligands are largely conserved (Heinisch et al., 1989; Deng et al., 1998; Rodicio et al., 2000). The high degree of similarity of these sites allowed us to identify the residues of these catalytically important amino acids. The two aspartic acid residues (Figure 2) found within the TIDGD sequence (residues 246-250) are highly conserved in the active site in all members of this superfamily of PPi-dependent PFKs (Deng et al., 1998). In addition, the sequence comparison of PPi-PFKs also shows that the arginine found at position 412 (Figure 2) is an essential binding site for the fructose 6-phosphate in all PPi-PFKs. The other sequence that is highly conserved in all PPi-PKFs is GGDD (Figure 2), found at position 216-220 (Arvanitidis and Heinisch, 1994; Raben et al., 1995; Deng et al., 1998). Another interesting finding of this sequence was the absence of the sequence GGED, suggesting that ATP is not an allosteric inhibitor of this enzyme. In addition, the differentiated expression of the enzyme’s a and b subunits in the different tissues of sugar cane (see Figure 1) may be an important physiological mechanism for the modulation and regulation of PPi-PFK during the process of in vivo sugar synthesis and storage. A similar argument has been used by Theodorou and Plaxton (1996), when they analyzed the expression of the enzyme’s a and b subunits in Brassica nigra. From the evidence that the enzyme’s a and b subunits may be present both in photosynthesizing and non-photosynthesizing tissues in sugar cane plants (Figure 1) and from the previous experimental results suggesting PPi-PFP activity in the sugar cane internodes (Lingle and Smith, 1991), it appears that this enzyme regulates the most critical branching point of sugar phosphate metabolism. The role of PPi-PFP appears to be to obtain energy from the PPi bond (the supposed cellular by-products), giving a considerable gain of energetic yield compared to the classical glycolysis model (Plaxton, 1996). In addition, PPi-PFK may be involved in important metabolic adaptations by which sugar cane can overcome environmental stresses during seasonal development. Though we now know that PPi-PFK is in sugar cane, and we have information about the location of enzyme ligands and of critical amino acid residues involved in its active site, we still do not know if these enzymes are expressed in vivo. Work in this direction is now in progress in our laboratory. ACKNOWLEGDMENTS Research supported by FAPESP. REFERENCES Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. Arvanitidis, A. and Heinisch, J.J. (1994). Studies on the function of yeast phosphofructokinase subunits by in vitro mutagenesis. J. Biol. Chem. 269: 8911-8918. Braam, J., Sistrunk, M.L., Polisensky, D.H., Xu, W., Purugganan, M.M., Antosiewicz, D.M., Campbell, P. and Johson, K.A. (1997). Plant responses to environmental stress: regulation and functions of the arabidopsis TCH genes. Planta 203: 535-541. Carlisle, S.M., Blakeley, S.D., Hemmingsen, S.M., Trevanion, S.J., Hiyoshi, T., Kruger, N.J. and Dennis, D.T. (1990). Conservation of protein sequence between the a- and b-subunits and with the ATP-dependent phosphofructokinase. J. Biol. Chem. 265: 18366-18371. Cheng, H.-F. and Tao, M. (1990). Differential proteolysis of the subunits of pyrophosphate-dependent 6-phosphofructo-1-phosphotransferase. J. Biol. Chem. 265: 2173-2177. Chi, A.S., Deng, Z., Albach, R.A. and Kemp, R.G. (2001). The two phosphofructokinase gene products of Entamoeba histolytica. J. Biol. Chem. 276: 19974-19981. Deng, Z., Huang, M., Singh, K., Albach, R.A., Latshaw, S.P., Chang, K.-P. and Kemp, R.G. (1998). Cloning and expression of the gene for the active PPi-dependent phosphofructokinase of Entamoeba histolytica. Biochem. J. 329: 659-664. Farré, E.M., Geigenberger, P., Willmitzer, L. and Trethewey, R.N. (2000). A possible role for pyrophosphate in the coordination of cytosolic and plastidial carbon metabolism within the potato tuber. Plant Physiol. 123: 681-688. Gibson, S.I. (2000). Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol. 124: 1532-1539. Gibson, S.I. and Graham, I.A. (1999). Another player joins the complex field of sugar-regulated gene expression in plants. Proc. Natl. Acad. Sci. USA 96: 4746-4748. Heinisch, J., Ritzel, R.G., von Borstel, R.C., Aguilera, A., Rodicio, R. and Zimmermann, F.K. (1989). The phosphofructokinase genes of yeast evolved from two duplication events. Gene 78: 309-321. Heldt, H.W. (1997). Polysaccharides. In: Plant Biochemistry & Molecular Biology. Oxford University Press Inc., New York, NY, USA, pp. 219-246. Ikeda, Y., Koizumi, N., Kusano, T. and Sano, H. (1999). Sucrose and cytokinin modulation of WPK4, an gene encoding an SNF1-related protein kinase from wheat. Plant Physiol. 121: 813-820. Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 509-540. Kruger, N.J. and Dennis, D.T. (1987). Molecular properties of pyrophosphate: fructose 6-phosphatase phosphotransferase from potato tuber. Arch. Biochem. Biophys. 256: 173-179. Lingle, S.E. and Smith, R.C. (1991). Sucrose metabolism related to growth and ripening in sugarcane internodes. Crop Sci. 31: 172-177. Nadkarni, M., Parmar, L., Lobo, Z. and Maitra, P.K. (1984). Mutations in the regulatory subunit of soluble phosphofructokinase from yeast. FEBS Lett. 175: 294-298. Nielsen, T.H. (1995). Fructose-1,6-biphosphate is an allosteric activator of pyrophosphate: fructose-6-phosphate 1-phosphotransferase. Plant Physiol. 108: 69-73. Plaxton, W.C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 185-214. Raben, N., Exelbert, R., Spiegel, R., Sherman, J.B., Nakajima, H., Plotz, P. and Heinisch, J. (1995). Functional expression of human mutant phosphofructokinase in yeast: genetic defects in French Canadian and Swiss patients with phosphofructokinase deficiency. Am. J. Hum. Genet. 56: 131-141. Rodicio, R., Straub, A. and Heinisch, J.J. (2000). Single point mutations in either gene encoding the subunits of the heterooctameric yeast phosphofrutokinase abolish allosteric inhibition by ATP. J. Biol. Chem. 275: 40952-40960. Sattler, R. and Rutishauser, R. (1997). The fundamental relevance of morphology and morphogenesis to plant research. Ann. Bot. 80: 571-582. Sheen, J., Zhou, L. and Jang, J.C. (1999). Sugars as signalling molecules. Curr. Opin. Plant Biol. 2: 410-418. Smeekens, S. (1998). Sugar regulation of gene expression in plants. Curr. Opin. Plant Biol. 1: 230-234. Stit, M. (1990). Fructose-2,6-biphosphate as a regulatory molecule in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 41: 153-185. Theodorou, M.E. and Plaxton, W.C. (1996). Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved Brassica nigra suspension cells. Plant Physiol. 112: 343-351. Wang, Y.H. and Shi, J.N. (1999). Evidence that fructose 1,6-bisphosphate specifically protect alpha-subunit of pyrophosphate-dependent 6-phosphofructo-phosphotransferase against proteolytic degradation. FEBS Lett. 459: 448-452. Yan , T.F. and Tao, M. (1984). Multiple forms of pyrophosphate: D-fructose-6-phosphate 1-phosphotransferase from wheat seedlings. Regulation by fructose 2,6-bisphosphate. J. Biol. Chem. 259: 5087-5092. Yu, S.M. (1999). Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 121: 687-693. |
|