
ABSTRACT. The complete genome sequence of the free-living bacterium Chromobacterium violaceum has been determined by a consortium of laboratories in Brazil. Almost 500 open reading frames (ORFs) coding for transport-related membrane proteins were identified in C. violaceum, which represents 11% of all genes found. The main class of transporter proteins is the primary active transporters (212 ORFs), followed by electrochemical potential-driven transporters (154 ORFs) and channels/pores (62 ORFs). Other classes (61 ORFs) include group translocators, transport electron carriers, accessory factors, and incompletely characterized systems. Therefore, all major categories of transport-related membrane proteins currently recognized in the Transport Protein Database (http://tcdb.ucsd.edu/tcdb) are present in C. violaceum. The complex apparatus of transporters of C. violaceum is certainly an important factor that makes this bacterium a dominant microorganism in a variety of ecosystems in tropical and subtropical regions. From a biotechnological point of view, the most important finding is the transporters of heavy metals, which could lead to the exploitation of C. violaceum for bioremediation. Key words: Genome, Bacterium, Transporters, Membrane, Biotechnology INTRODUCTION Chromobacterium violaceum is a Gram-negative, b-proteobacterium; it is a dominant microorganism in diverse ecosystems in tropical and subtropical regions, thus providing an excellent model for the study of environmental adaptation strategies. Recently, the complete genome sequence of C. violaceum type strain ATCC 12472 (Vasconcelos et al., 2003) has been determined and annotated by the Brazilian National Genome Sequencing Consortium (www.brgene.lncc.br). Transport-related membrane proteins mediate this bacterium’s direct metabolic interactions with the complex soil and aquatic environments that it inhabits. Therefore, the analysis of C. violaceum transport proteins is a key step to unravel the strategies evolved by this microorganism to adapt to complex environmental conditions. We present an overview of the transport capabilities encoded in the C. violaceum genome. NUTRIENT AND ION TRANSPORTERS Transport systems allow the uptake of essential nutrients and ions, excretion of end products of metabolism and of deleterious substances, and communication between cells and the environment (Pao et al., 1998). The physiology of transport systems mediating the uptake of nutrients by bacteria (particularly E. coli and Salmonella typhimurium) was studied in detail in the 1970s. It soon became apparent that bacteria had multiple systems for the uptake of most nutrients and that these systems fell into a small number of classes. Currently, seven major categories are recognized according to the Transport Protein Database (TCDB): channels and pores (class 1), electrochemical potential-driven transporters (class 2), primary active transporters (class 3), group translocators (class 4), transport electron carriers (class 5), accessory factors involved in transport (class 6), and incompletely characterized transport systems (class 7). The major facilitator superfamily (MFS) is a well-known transport family that belongs to the secondary, shock-insensitive transporters that are energized by the electrochemical gradient, while ABC transporters are an important family in the primary, shock-sensitive system energized directly by the hydrolysis of ATP (Saier, 2000; Higgins, 2001). The complete genome of C. violaceum comprises a single circular chromosome of 4,751,080 bp containing 4,431 open reading frames (ORFs). According to TCDB criteria, about 12.2% of all annotated ORFs (known + conserved hypothetical + hypothetical) in C. violaceum genome 539 ORFs encode proteins involved in the transport of metabolites. Most of these ORFs (489, which represents 11% of all annotated ORFs) encode protein sequences that have significant similarities to transport-related membrane proteins from other organisms (Table 1). The remaining 50 ORFs encode protein sequences comprising conserved hypothetical and hypothetical proteins that have been classified as transporters by TCDB, although they do not show any significant sequence similarities to known proteins from other organisms. The significant number of potential transporters in C. violaceum may be related to the fact that this microorganism lives in diverse environments in tropical and subtropical regions and needs to adapt to a great array of external conditions. 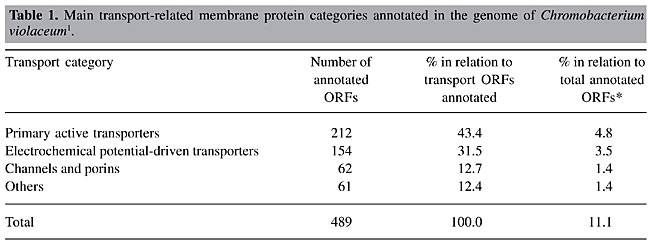
1www.brgene.lncc.br/cviolaceum
*valid ORFs + conserved hypothetical ORFs + hypothetical ORFs = 4,431. General diffusion Gram-negative channels and porins The outer membrane of Gram-negative bacteria acts as a molecular filter for hydrophilic compounds. Proteins, known as porins, are responsible for the ‘molecular sieve’ properties of the outer membrane. Porins form large water-filled channels, which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solutes up to a certain size (that size is known as the exclusion limit) to cross the membrane, while other porins are specific for a solute and contain a binding site for that solute inside the pores (these are known as selective porins). As porins are the main outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins (Benz and Bauer, 1988). General diffusion porins generally assemble as trimers in the membrane, and the transmembrane core of these proteins is composed exclusively of beta strands (Jap and Walian, 1990). A number of general porins are evolutionary related; these porins are: phoE, ompC, ompF, and nmpC (Jeanteur et al., 1991). Channels and porins (62 ORFs) comprise the third-most numerous class of transporters in C. violaceum (Table 1), including 17 a-type channels and 41 b-barrel porins. Among the channel/porin proteins, there is one ORF coding a member of the large conductance mechanosensitive channel (MscL) and two ORFs coding members of the small conductance mechanosensitive channel (MscS). The sensing of physical forces within a cell’s environment is primarily mediated by this specialized class of membrane proteins, known as mechanosensitive ion channels. Mechanosensitive channels have evolved the ability to transduce mechanical stress into an electrochemical response (Sackin, 1995) enabling cells to respond to stimuli such as sound, touch, gravity, and pressure. The major facilitator superfamily Electrochemical potential driven transporters (154 ORFs) are the second-most abundant group of transporters in C. violaceum, accounting for 31.5% of all annotated ORFs related to transport (Table 1). Within this category, the MFS utilizes an electrochemical ion gradient to facilitate solute transport. This is a widespread grouping of secondary transporters, containing a single subunit with 12 membrane-spanning helices, for example, lactose permease from E. coli (LacY) (Pao et al., 1998). MFS transporters comprise most (about 29.9%) of the electrochemical potential driven transporters found in C. violaceum. Most of these MFS proteins are general substrate transporters or are related to multidrug resistance. In addition to the MFS transporters, electrochemical potential driven transporters involved in nutrient uptake belonging to the following families have been annotated (the number of genes in each transport family is shown in parentheses): concentrative nucleoside transporter, CNT (1), proton-dependent oligopeptide transporter, POT (2), cation diffusion facilitator, CDF (2), C4-dicarboxylate uptake, dcu (2), inorganic phosphate transporter, PiT (1), hydroxy/aromatic amino acid permease, HAAAP (1), formate-nitrite transporter, FNT (1), ammonium transporter, Amt (1), amino acid-polyamine-choline, APC (8), K+ uptake permease, KUP (2), Ca2+:cation antiporter, CaCA (1), betaine/carnitine/choline transporter, BCCT (1), monovalent cation:proton antiporter-2, CPA2 (3), dicarboxylate/amino acid:cation (Na+ or H+) symporter, DAACS (3), alanine/glycine:cation symporter, AGCS (3), lactate permease, LctP (1), nucleobase:cation symporter-1, NCS1 (1), nucleobase:cation symporter-2, NCS2 (2), solute:sodium symporter, SSS (2), neurotransmitter:sodium symporter, NSS (3), citrate:cation symporter, CCS (1), glutamate:Na+ symporter, ESS (1), NhaC Na+:H+ antiporter, NhaC (1), and glycerol uptake, GUP (1). Putative transporters for trace elements (arsenic, cadmium, cobalt, copper, chromium, lead, mercury, nickel, zinc) are also present in C. violaceum. These elements are found at low concentrations in rocks, soil, water, and the atmosphere; however, at high concentration they are toxic to organisms (Madigan et al., 2002). Some of the microbial proteins that transport heavy metals are involved in bacterial resistance to them. Therefore, C. violaceum has a potential to be genetically modified with key catabolic genes, which could make it useful for environmental remediation. ABC transporters The ABC (ATP-binding cassette) protein transporters are the second major family of solute transport systems encoded in prokaryote genomes. The ABC transporters contain a cytoplasmic domain (the ABC protein) that binds and hydrolyses ATP to energize solute translocation across the cytoplasmic membrane. The number of ABC transporters differs widely between species. Organisms such as E. coli, which live in diverse environments and need to adapt to a great array of external conditions, have many of these transporters; the E. coli chromosome encodes around 70 ABC transporters. In contrast, some other species have far fewer examples, perhaps reflecting their more restrictive lifestyles (Saier, 2000; Higgins, 2001). Although, in general, each ABC transporter is relatively specific for its own particular substrate(s), there is an ABC transporter for essentially every type of molecule that must cross a cellular membrane. ABC transporters have been characterized with specificity for small molecules, large molecules, highly charged molecules, and highly hydrophobic molecules; systems are known with specificity for inorganic ions, sugars, amino acids, proteins, and complex polysaccharides. Although most exhibit relatively tight substrate specificity, some are multispecific, such as the oligopeptide transporter, which can handle essentially all di- and tripeptides (Madigan et al., 2002). The basic unit of an ABC transporter consists of four core domains. Frequently, each of the four core domains is encoded as a separate polypeptide (e.g., the oligopeptide transporter), although in other transporters the domains can be fused in any one of a number of ways into multidomain polypeptides. In cases in which one of the four domains is absent, one of the remaining domains functions as a homodimer to maintain the full complement. The two transmembrane domains span the membrane multiple times via putative a-helices. Typically, there are six predicted membrane-spanning a-helices per domain (a total of 12 per transporter), although there is some variation on this formula. The transmembrane domains form the pathway through which solute crosses the membrane, and they determine the specificity of the transporter through substrate-binding sites. The other two domains, the ATP or nucleotide-binding domains, are hydrophilic and peripherally associated with the cytoplasmic face of the membrane. These domains consist of the core 215 or so amino acids of the ABC domain by which these transporters are defined (Saier, 2000; Higgins, 2001). In many ABC transporters, auxiliary domains have been recruited for specific functions. The periplasmic binding proteins bind substrates external to the cell and deliver them to the membrane-associated transport complex. It appears that periplasmic binding proteins have two distinct but related functions: a) the first is to impart high affinity and specificity; b) the second is to confer directionality. Other ABC transporters require outer membrane proteins to facilitate solute entry into the periplasm (e.g., transporters for iron chelates), while Gram-negative ABC transporters, which mediate protein export, require additional outer membrane proteins to facilitate transport across the periplasm and outer membrane (e.g., the HlyD and TolC proteins required for export of hemolysin) (reviewed by Higgins, 2001). The largest group of ORFs (212) coding for transport-related membrane proteins in C. violaceum genome (Table 1) belongs to the class of primary active transporters (43.3% of the putative transporters), of which 119 are ABC type. Indeed ABC-type transporters constitute the main transport system in C. violaceum (24.3% of all transport-related ORFs). Furthermore, this transport system includes about 2.7% of the annotated ORFs in the C. violaceum genome. Most (79.4%) of the ABC-type transport ORFs in C. violaceum are dedicated to nutrient acquisition, while the remaining are related to multidrug resistance. The molecules/metabolites transported by ABC-type transport systems in C. violaceum are as follows: phosphates/phosphonates, sulfate/molybdate, amino acids (glutamate/aspartate, leucine/isoleucine/valine, arginine/ornithine, taurine, histidine), metals (copper, iron, magnesium, manganese/zinc), nitrate/nitrite, spermidine/putrescine, dipeptide/oligopeptide/nickel, potassium (K+) and sugars (ribose, xylose, arabinose, galactose, maltose and glycerol-3-phosphate). The great majority of these systems are organized in gene clusters (which probably work as operons) comprising three components: an ATP-binding protein/transporter (the ATPase component) and two auxiliary proteins, a permease and a substrate-binding protein (usually located in the periplasmic space). This means that these ABC-type transporters are designed to transport nutrients with the following characteristics: high affinity and specificity, directionality and facility. In E. coli, Vibrio cholerae and other Gram-negative bacteria, OmpF-type outer membrane porins allow the passive diffusion of hydrophilic substrates across the outer membrane. Only three genes coding for OmpA-OmpF-type porins have been annotated (CV3571, CV0110, CV1891) in the C. violaceum genome. Therefore, the relative abundance of active, ATP-binding cassette domain transporters, which usually have high affinity for their substrates, may be essential for C. violaceum nutrient uptake and survival in its habitat. Caulobacter crescentus, a Gram-negative, free-living bacterium that grows in dilute aquatic environments, has developed another strategy to acquire nutrients in nutrient-limiting conditions. It has 65 members of the family of TonB-dependent outer membrane channels that catalyze energy-dependent transport across the outer membrane (Nierman et al., 2001). Only six of such TonB-dependent receptors have been annotated (CV0077, CV1019, CV1699, CV1970, CV3188, and CV3896) in the C. violaceum genome. Usually, the proteobacteria sequences have no more than 10 of these transporters (Nierman et al., 2001). Primary active transporters drive solute accumulation or extrusion, by using ATP hydrolysis, photon absorption, electron flow, substrate decarboxylation, or methyl transfer. The ABC family of transporters uses ATP hydrolysis to energize solute translocation across the cytoplasmic membrane. If charged molecules are unidirectionally pumped, as a consequence of the primary consumption of a primary cellular energy source, electrochemical potentials result. The consequential chemiosmotic energy that is generated can then be used to drive the active transport of additional solutes via secondary carriers, which merely facilitate the transport of one or more molecular species across the membrane (Pao et al., 1998). These are the so-called electrochemical-potential-driven transporters. The fact that these transporters are the second largest group of transport proteins in C. violaceum could be putatively viewed as an efficient way that this organism has found to take advantage of the electrochemical potentials that result from the activity of the most abundant ABC-type transporters. Iron transport (uptake): regulation, and its relation to pathogenesis Iron is the fourth most abundant metal on Earth; however, it is found in the environment as a component of insoluble hydroxides, and in biological systems it is chelated by high-affinity iron binding proteins or is present as a component of erythrocytes. Iron acquisition is an essential requirement for all microorganisms, except certain lactobacilli and Borrelia burgdorferi. It is essential, because it is a component of key molecules, such as cytochromes, ribotide reductase, and other compounds involved with metabolism. However, iron can also be deleterious: hydroxyl free radicals generated through Haber-Weiss reactions catalyzed by iron accumulate, leading ultimately to cell death. Consequently, the production of the cellular components responsible for utilizing iron is controlled by various parameters that act under different physiological and environmental conditions in either a negative (under iron-rich conditions) or a positive (under iron-limiting conditions) fashion (Crosa, 1997; Koster, 2001). One important control comes directly from iron itself. High concentrations of this metal leads to a shut-off of the expression of many genes involved in iron uptake; this occurs in conjunction with the Fur protein, which acts as a repressor, together with iron (reviewed by Crosa, 1997). Fur is the product of the fur (ferric uptake regulation) gene, which controls the transcription of iron-dependent promoters in many prokaryotes. This regulator is a zinc-containing, Fe2+-binding protein that inhibits the transcription of genes implicated in the response to iron starvation when the metal is in excess in the medium. But Fur also appears to play an important role in a variety of cell functions unrelated to iron acquisition, such as the production of several virulence determinants, defense against oxygen radicals, the acid shock response, chemotaxis, and metabolic pathways (Crosa, 1997). The interaction of the Fur protein-Fe2+ complex with its operators has been characterized with diverse techniques in several promoters of E. coli and other genera. These studies have revealed that every iron-dependent promoter contains a target DNA sequence with different degrees of similarity to a palindromic 5’-GATAATGATAATCATTATC-3’, 19-bp consensus box. More recently, such a consensus has been reinterpreted as the combination of three repeats of the simpler motif 5’-NAT(A/T)AT-3’, in which the thymines are the bases determining the type of contact of the Fur protein with such a minimal unit of interaction. The corollary of this interpretation is that extended sites for Fur binding could be naturally or artificially assembled by simply adding multiple adjacent 5’-NAT(A/T)AT-3’ hexamers to a minimum of three repeats. This is a very attractive possibility, because it would permit the generation of repertoires of binding sites of varying extensions and affinities, which would allow Fur to act in some promoters as a very specific regulator and in others as a more general co-regulator (Crosa, 1997; Hantash and Earhart, 2000). The Fur protein in C. violaceum is encoded in ORF CV1797. It would be interesting to determine the Fur binding DNA consensus box in C. violaceum gene promoters. This would give an idea about which genes have their expression affected by iron availability. As iron uptake plays an important role in pathogenicity, this information could give some insight about pathogenicity in C. violaceum. The abilities of bacterial pathogens to adapt to the environment within the host are essential to their virulence. In mammals, iron is bound to eukaryotic proteins (hemoglobin, ferritin, transferrin and lactoferrin), which maintain a level of free iron much too low (10-18 M) to sustain bacterial growth. In response to infection, the availability of free iron in body fluids is further reduced by shifting iron from transferrin to lactoferrin in the liver. This process is called induced hypoferremia and forms part of the non-specific immune response (Carniel, 2001). Microorganisms have adapted to the iron limitation present in mammalian hosts by developing diverse mechanisms for the assimilation of sufficient iron for growth. In addition, many bacterial pathogens have used the low concentration of iron present in the host as an important signal to enhance the expression of a wide variety of bacterial toxins and other virulence determinants. Siderophores and iron uptake One of the most widely used solutions employed by bacteria to acquire iron in an iron-restricted environment is the synthesis and secretion of low-molecular mass Fe3+-chelating compounds, designated siderophores, which may be regarded as virulence factors. Because of their high affinity for iron, siderophores can solubilize the metal bound to host binding proteins and transport it into the bacteria. The siderophore-Fe3+ complex recognizes a specific bacterial outer membrane receptor, and it is translocated into the cytosol with the help of proteins located in the periplasm and the inner membrane of the cell wall. The iron is discharged from its siderophore in the bacterial cytosol, and it is utilized for different metabolic pathways (reviewed by Koster, 2001). Three major structural types that are involved in complexing ferric iron are: catecholates, hydroxamates, and a-hydroxycarboxylates. Transport of the enterobactin-iron complex Escherichia coli responds to iron deprivation by synthesizing and excreting a small, iron-chelating molecule, termed enterobactin (Ent), which is a cyclic trimeric lactone of N-(2,3-dihydroxybenzoyl)serine. Enterobactin is a tricatecholate compound that has an extremely high affinity for Fe3+. It captures exogenous Fe3+ by forming a complete six-ligand coordination sphere around the iron. Complexes of extracellular Fe(III)-Ent are subsequently transported into the cytoplasm, where Fe(III) is reduced and released from enterobactin (Koster, 2001). The enterobactin biosynthetic pathway is known. It has two stages; in the first, chorismate is converted to the specific precursor, 2,3-dihydroxybenzoate (DHBA) by the EntC, -B/G and -A proteins, and in the second, three molecules each of DHBA and Ser are converted by EntB/G, -D, -E, and -F to Ent, by a protein-thiotemplate mechanism. EntD, a phosphopantetheinyl transferase, post-translationally modifies both EntB/G and EntF by adding 4’-phosphopantetheine. Holo-EntB/G, holo-EntF, and Ent-E (collectively termed Ent synthase) then catalyze Ent formation (reviewed by Hantash and Earhart, 2000). Genes with similarity to EntA (CV1482), EntB (CV1483), EntC (CV1485), EntD (CV2650), EntE (CV1484), and EntF (CV1486) have been annotated in the C. violaceum genome. Fe-Ent is specifically recognized by FepA, an E. coli outer membrane protein that imports Fe-Ent into the periplasm. It is subsequently bound by a periplasmic binding protein, FepB, and transported across the inner membrane by the FepCDG complex. The FepCDG complex (FepC, FepD, FepG) is an ATP-dependent ABC transporter, but the overall process is limited by the ability of FepA to carry out active transport with the assistance of inner membrane proteins TonB and ExbBD (ExbB, ExbD) and the inner membrane chemiosmotic proton gradient (Usher et al., 2001). All these genes have been identified in the C. violaceum genome: FepA (CV2230), FepB (CV2239), FepC (CV2234), FepD (CV2236), FepG (CV2235), TonB (CV4254), ExbB (CV0399 and CV3348), and ExbD (CV0398, CV1973, CV1974, CV1985, CV1986, and CV3347). Once inside the E. coli cell, the iron is released from the enterobactin-iron complex by the enzyme enterochelin esterase, encoded by fes. The protein Fes is also able to degrade the free enterobactin (Greenwood and Luke, 1978). The fes gene has been found in the C. violaceum genome (CV2231). Therefore, it seems that C. violaceum is able to synthesize enterobactin-like siderophores, transport the enterobactin-Fe3+ complex back into the cell, release the iron from the enterobactin-Fe3+ complex, and degrade the siderophore. Transport of the ferrichrome-iron complex Ferrichrome is an iron complex of the hydroxamate type siderophores. FhuA is an E. coli outer membrane protein, which transports the ferric siderophore ferrichrome, together with the TonB-ExbB-ExbD protein complex, in the cytoplasmic membrane. The siderophore transport activity of the outer membrane protein FhuA of E. coli requires the proton motive force of the cytoplasmic membrane. It is postulated that the energy of the proton motive force is transduced to the transport proteins by a protein complex that consists of the TonB, ExbB, and ExbD proteins (the same mechanism used for enterobactin-iron uptake). The ferrichrome is subsequently transported across the inner membrane by the FhuBCD complex, an ABC-type transport system (Koster, 2001). The following proteins have been identified in the C. violaceum genome: FhuA (CV2251); TonB (CV4254); ExbB (CV0399 and CV3348); ExbD (CV0398, CV1973, CV1974, CV1985, CV1986, and CV3347), and FhuC (CV1487, CV1560, and CV1793). Two of the ORFs coding FhuC (CV1487 and CV1560) are located in clusters (which probably work as operons), each one containing two other ORFs: one coding for an ABC-type iron permease (CV1488 and CV1558, respectively) and the other coding an ABC-type iron substrate binding component (CV1489 and CV1559, respectively). In the FhuBCD ABC-type iron transport system, FhuD is the substrate-binding, periplasmic component of the transporter, while FhuB is the permease component (Koster, 2001). If we assume that the proteins encoded by these ORFs and clustered together with C. violaceum FhuC are the equivalents of FhuB and FhuD, then C. violaceum would be able to transport (uptake) the hydroxamate type siderophore ferrichrome. Iron uptake without siderophores: ABC transporters of the ferric iron type They are thought to mediate the further transport into the cytoplasm of ferric iron that is acquired from lactoferrin or transferrin and delivered into the periplasm in a receptor-mediated Ton complex-dependent fashion. A rather uniform organization of ferric iron transport genes has been observed in most bacteria studied so far: a putative iron-regulated operon containing genes encoding the substrate-binding protein, the permease component, and the ATPase, in that order. In addition, the ferric iron-type ABC-transport proteins display a low but significant degree of homology to the equivalent components that are involved in the utilization of substances such as sulfate, spermidine and putrescine (Koster, 2001). Based on these characteristics, we could identify five putative ABC transport systems of the ferric iron type in the C. violaceum genome, which are denoted here as clusters I, II, III, IV, and V. Four of them (I, III, IV, and V) display the predicted organization as mentioned above. Furthermore, two clusters (I and V) have two permease components in the operon, and cluster IV is unusual since it has, besides the genes encoding the three ABC protein components, a fourth one encoding a putative acetyltransferase. Iron uptake without siderophores: ABC transporters of the metal type The substrate-binding proteins of this type of system were originally described as adhesions in a variety of streptococcal pathogens. Transport systems of the metal type are established in many species. Not all of them are primarily involved in the acquisition of iron: some have a higher specificity for other metals, such as zinc and manganese; for others it has been clearly shown that they are essential for iron acquisition (e.g., Yfe of Yersinia pestis and Sit of Salmonella typhimurium) (Koster, 2001). A cluster of three genes encoding an ABC transporter of the metal type was identified in the C. violaceum genome: CV3064, encoding a periplasmic Mn2+/Zn2+-binding (lipo)protein (surface adhesin A), CV3065, encoding a Mn2+/Zn2+ permease component, and CV3066, encoding the ATPase component. In addition, a second copy of the gene encoding a putative periplasmic Mn2+/Zn2+-binding (lipo)protein (surface adhesin A) is also present (CV1154). The description of the substrate-binding proteins (encoded by CV3064 and CV1154) as surface adhesin A can be taken as evidence that this system is involved in iron acquisition, as has been demonstrated for Yfe and Sit. In addition, the components of the C. violaceum transport system show similarity with the corresponding components of SitABCD of S. typhimurium, which is an iron-transporter system. In this operon, SitA protein is a periplasmic binding protein, SitB encodes an ATP-binding protein, and SitC and SitD encode two putative permeases (integral membrane proteins) (Zhou et al., 1999; Janakiraman and Slauch, 2000). Iron uptake without siderophores: the Nramp system The natural resistance-associated macrophage protein (NRAMP) family consists of Nramp1, Nramp2, and yeast proteins Smf1 and Smf2. The NRAMP family is a novel family of functionally related proteins, defined by a conserved hydrophobic core of 10 transmembrane domains (Cellier et al., 1995). Nramp1 is an integral membrane protein expressed exclusively in cells of the immune system, and it is recruited to the membrane of a phagosome upon phagocytosis. Nramp2 is a multiple divalent cation transporter for Fe2+, Mn2+ and Zn2+, amongst others. It is expressed at high levels in the intestine, and is a major transferrin-independent iron uptake system in mammals (Govoni and Gros, 1998). The yeast proteins Smf1 and Smf2 may also transport divalent cations (Agranoff and Krishna, 1998). Three C. violaceum ORFs encode proteins classified as belonging to the NRAMP family of Mn2+ and Fe2+ electrochemical driven transporters: CV3478, CV3314 and CV0576. Iron storage Besides been able to employ different systems to acquire iron from the environment, C. violaceum also has an efficient mechanism to store the transported metal. This is done by two proteins: bacterioferritin (BFR) (two genes were found, CV3399 and CV3552) and a frataxin-like homolog (CV0040). The BFR (also known as cytochrome b1 or cytochrome b557) (Andrews et al., 1990, 1991) of E. coli is an iron-storage protein, consisting of 24 identical subunits that pack together to form a highly symmetrical, nearly spherical shell, surrounding a central cavity about 8 nm in diameter (Le Brun et al., 1995; Harrison and Arosio, 1996). X-ray crystallographic studies have revealed a close structural similarity between BFR and the ferritins, a family of iron-storage proteins found in both eukaryota and prokaryota (Harrison and Arosio, 1996). Common to both ferritins and BFRs is a capacity to store large quantities of iron within their hollow interior, in the form of a hydrated ferric oxide mineral containing variable amounts of phosphate anion. However, a major difference between them is that BFR contains up 12 b-type haem groups, while ferritins, when isolated, do not contain haem. The building block for the BFR shell is a protein dimer (subunits A and B), binding the single haem group. Each subunit consists of four nearly parallel alpha-helices. The haem is bound symmetrically to subunits A and B by Met(A)-52 and Met(B)-52 residues (Frolow et al., 1994). Each subunit includes a binuclear metal-binding site, linking together the four major helices of the subunit, which has been identified as the ferroxidase center of BFR (Le Brun et al., 1995). BFR mutants with Met-52 replaced are haem-free, but appear to be correctly assembled and are capable of accumulating iron (Andrews et al., 1995). Another protein that might play an important role in iron storage is the frataxin homolog (named cya Y protein), which has been implicated in iron transport. The frataxin-like domain is related to the globular C-terminus of frataxin, the protein that is mutated in Friedreich’s ataxia (Gibson et al., 1996). Friedreich’s ataxia is a progressive neurodegenerative disorder caused by loss of function mutations in the gene encoding frataxin. Frataxin mRNA is predominantly expressed in tissues with a high metabolic rate (including liver, kidney, brown fat, and heart muscle). Mouse and yeast frataxin homologues contain a potential N-terminal mitochondrial targeting sequence, and human frataxin has been observed to co-localize with a mitochondrial protein. Furthermore, disruption of the yeast gene has been shown to result in mitochondrial dysfunction. Friedreich’s ataxia is thus believed to be a mitochondrial disease caused by a mutation in the nuclear genome (specifically, expansion of an intronic GAA triplet repeat) (Campuzano et al., 1996; Durr et al., 1996; Koutnikova et al., 1997). This domain is found in a family of bacterial proteins. However, its function is currently unknown. Recently, it has been shown that when expressed in E. coli, the mature form of human frataxin assembles into a stable homopolymer that can bind approximately 10 atoms of iron per molecule of frataxin. Moreover, in radio-labeled yeast cells, human frataxin is recovered by immunoprecipitation with approximately five atoms of (55)Fe bound per molecule (Cavadini et al., 2002). The authors thus suggest that Friedreich’s ataxia results from decreased mitochondrial iron storage due to frataxin deficiency, which may impair iron metabolism, promote oxidative damage and lead to progressive iron accumulation. Therefore, it can be speculated that the frataxin homolog present in C. violaceum and other bacteria can also work as an iron storage protein. In conclusion, C. violaceum seems to have the ability to acquire iron very efficiently by using different systems of iron transport: siderophore (enterobactin)-mediated uptake, ferrichrome-mediated uptake (to be confirmed), ABC-type transport of the metal type, ABC-type transport of the ferric ion type and electrochemical driven transporters of the NRAMP family. Upon acquisition, the metal is stored by using BFR and the frataxin homolog protein. Iron uptake and bacterial pathogenesis The important role that iron uptake plays in bacterial pathogenesis has been demonstrated for various species. For instance, highly pathogenic Yersinia carries a pathogenicity island termed high-pathogenicity island (HPI). The Yersinia HPI comprises genes involved in the synthesis of the siderophore yersiniabactin, and can thus be regarded as an iron-uptake island. Pathogenic Yersinia can be further subdivided into low-pathogenicity strains, i.e., strains that induce a mild intestinal infection in humans and are non-lethal for mice at low doses, and high-pathogenicity strains, which cause severe systemic infections in humans and are lethal to mice at low doses. Although other, as yet unidentified, factors may participate in the high-pathogenicity phenotype, one of the major differences between low- and high-pathogenic Yersinia lies in the ability of the latter to capture the iron molecules necessary for their systemic dissemination in the host. Investigation of large numbers of different Yersinia strains indicates that the presence of HPI-specific genes or products correlates with their level of pathogenicity. Furthermore, HPI has never been detected in low-pathogenic or avirulent strains of Yersinia (reviewed by Carniel, 2001). An iron uptake system encoded in the Salmonella typhimurium pathogenicity island-1 (SPI1) was identified and characterized by Zhou et al. (1999). This locus was designated sit (Salmonella iron transporter) and consists of four genes (sitABCD). This system belongs to the ABC family of transporters, with extensive homology with the yfe ABC iron transport system of Yersinia pestis. The sitA gene encodes a putative periplasmic binding protein, sitB encodes an ATP-binding protein, and sitC and sitD encode two putative permeases (integral membrane proteins). It has been demonstrated that sitABCD is required for full virulence of S. typhimurium (Janakiraman and Slauch, 2000). A cluster of 22 ORFs similar to SPI1 has been identified in the C. violaceum genome. An operon similar to the sitABCD, or otherwise involved in iron uptake, is not present in this region. However, there is a cluster of three ORFs encoding an ABC iron transport system of the metal type, whose components have some similarity with those from the sitABCD operon and from Yfe of Y. pestis, which is also an ABC type iron transport system involved in pathogenesis (see item “Iron uptake without siderophores: ABC transporters of the metal type”). DRUG TRANSPORTERS Microorganisms have developed various ways to resist the toxic effects of antibiotics and other drugs. One of these mechanisms involves the production of enzymes that inactivate antibiotics by hydrolysis or by the formation of inactive derivatives. A second mechanism of resistance is target alteration. Cellular targets can be altered by mutation or enzymatic modification in such a way that the affinity of the antibiotic for the target is reduced. A third, more general, mechanism of resistance is the inhibition of drug entry into the cell. Due to the low permeability of the outer membrane of Gram-negative bacteria and the exceptionally efficient barrier of the Gram-positive mycobacteria, drug diffusion across the cell envelope is reduced. The permeability of the outer membrane can be further decreased by the loss of porins. However, these barriers cannot prevent the drugs from exerting their toxic action once they have entered the cell, and the active efflux of drugs is essential to ensure significant levels of drug resistance (reviewed by Putman et al., 2000). Some transporters, such as the tetracycline efflux proteins, are dedicated systems, which mediate the extrusion of a given drug or class of drugs. In contrast to these specific drug transporters, the so-called multidrug transporters can handle a wide variety of structurally unrelated compounds. Multidrug transporters can be divided into two major classes on the basis of bioenergetic and structural criteria. Secondary multidrug transporters utilize the transmembrane electrochemical gradient of protons or sodium ions to drive the extrusion of drugs from the cell. ABC-type multidrug transporters use the free energy of ATP hydrolysis to pump drugs out of the cell (Pao et al., 1998; Putman et al., 2000; Saier, 2000). Secondary multidrug transporters This grouping comprises most bacterial multidrug efflux systems known to date. They mediate the extrusion of structurally unrelated drugs in a coupled exchange with protons (H+) or sodium (Na+) ions. These secondary multidrug transporters can be subdivided into distinct families of transport proteins on the basis of size and similarities in the primary and secondary structure: the MFS, the small multidrug resistance (SMR) family, the resistance-nodulation-cell division (RND) family, and the multidrug and toxic compound extrusion (MATE) family (Putman et al., 2000). The MFS consists of membrane transport proteins, which are found from bacteria to higher eukaryotes, and are involved in the symport, antiport, and uniport of various substrates, such as sugars, Krebs cycle intermediates, phosphate esters, oligosaccharides, and antibiotics. Hydropathy analysis and alignment of conserved motifs of the resistance-conferring drug efflux proteins revealed that these proteins can be divided into two separate clusters, with either 12 or 14 transmembrane segments. The multidrug transporters of the SMR family, the smallest secondary drug efflux proteins known, are typically about 107 amino acid residues in length. Due to the small size of the multidrug transporters of the SMR family, it has been proposed that they may function as homooligomeric complexes. Multidrug transporters belonging to the RND family interact with a membrane fusion protein and an outer membrane protein to allow drug transport across both the inner and outer membrane of Gram-negative bacteria. The membrane fusion proteins, which contain a single N-terminal transmembrane segments and a large C-terminal periplasmic domain, are thought to induce fusion of the inner and outer membrane, to form a channel-like structure that spans the periplasmic space (Pao et al., 1998; Putman et al., 2000). ATP-dependent multidrug transporters Although most bacterial multidrug transporters utilize the proton motive force (or sodium) for the extrusion of cytotoxic compounds, some drug efflux systems are driven by the free energy of ATP hydrolysis. All ATP-dependent drug efflux proteins known to date are members of the ABC superfamily, also referred to as traffic ATPases. Unlike the ABC type transporters involved in solute uptake, the ATP-dependent transporters used for drugs and antibiotics do not contain the auxiliary, extracytoplasmic solute receptor component (the periplasmic binding protein) that is required for solute transport (Saier, 2000; Higgins, 2001). Both major systems of active drug/multidrug transporters (secondary, electrochemical gradient-dependent and ATP-dependent) are present in the C. violaceum genome. Forty-six putative transport-related membrane proteins are involved in multidrug resistance, with a predominance of electrochemical potential driven transporters (70% of drug/multidrug transporters). The secondary multidrug transporters belong to the major facilitator superfamily (17), to the RND family (10 ORFs), to the MATE family (1), to the membrane fusion protein family (1) and to a group of unknown transporters (2). This multidrug resistance apparatus is thought to be an important characteristic that allows C. violaceum to withstand environmentally unfavorable conditions. PATHOGENESIS-RELATED TRANSPORTERS Gram-negative bacterial pathogens have evolved sophisticated mechanisms to infect and colonize their hosts. Some of these mechanisms require the assembly of multicomponent organelles on the bacterial surface. These organelles can be quite large and complex, consisting of many different proteins and hundreds of individual subunits. Prior to their assembly, each subunit must first be exported and localized to its point of incorporation within a growing structure. Because the cell envelope of Gram-negative bacteria presents several barriers to the movement of organellar components, bacteria have evolved unique protein secretion/transport mechanisms to facilitate surface organelle assembly. Organelles such as fimbriae utilize the general secretory pathway for secretion of components across the inner membrane, followed by unique and divergent mechanisms for secretion and assembly of fimbrial subunits beyond the outer membrane. In contrast, the type III secretion pathway, which functions in the assembly of both flagella and virulence-associated organelles, secretes proteins across both membranes, independently of the secretory pathway, without the need for a periplasmic intermediate or proteolytic processing (Kimbrough and Miller, 2002). Therefore, secretion of virulence factors in Gram-negative bacteria involves transportation of the protein across two membranes to reach the cell exterior (Mecsas and Strauss, 1996). There have been four secretion systems described in animal enteropathogens, such as Salmonella and Yersinia, with further sequence similarities in plant pathogens, including Ralstonia and Erwinia (Mecsas and Strauss, 1996). The type III secretion system (TTSS) is of great interest, as it is used to transport virulence factors from the pathogen directly into the host cell (Galan and Collmer, 1999), and it is only triggered when the bacterium comes into close contact with the host. The protein subunits of the TTSS are very similar to those found in bacterial flagellar biosynthesis (Komoriya et al., 1999). However, while the latter forms a ring structure to allow secretion of flagellin and is an integral part of the flagellum itself (Komoriya et al., 1999), type III subunits in the outer membrane translocate secreted proteins through a channel-like structure. It is believed that the family of type III inner membrane proteins is used as structural moieties in a complex with several other subunits (Hueck, 1998). One such set of inner membrane proteins, labeled here “S” for nomenclature purposes, includes the Salmonella and Shigella SpaS, the Yersinia YscU, Rhizobium Y4YO, and the Erwinia HrcU genes (Hueck, 1998). The flagellar protein FlhB also shares similarity, probably due to evolution of the TTSS from the flagellar biosynthetic pathway. Type III secretion system Virulence-associated TTSS are specialized organelles that translocate bacterial virulence proteins (effectors) from the bacterial cytoplasm directly into the host-cell cytoplasm. These translocated effectors alter such basic host-cell functions as signal transduction, cytoskeletal architecture, membrane trafficking, and cytokine gene expression. Many Gram-negative pathogens, such as Yersinia, Salmonella, Erwinia, and Pseudomonas require this system to cause disease in a number of animal and plant hosts. Although the translocated effectors vary between pathogens, recent visualization of virulence-associated TTSS organelles from Salmonella, Shigella, and Yersinia reveals that the organellar structure itself is well conserved. Perhaps the most thoroughly characterized virulence type III secretion organelle is the SPI1-encoded TTSS of Salmonella typhimurium, termed the needle complex. All the genes required for needle complex assembly and function are located on SPI1, a 40-kb gene cluster at centisome 63 of the S. typhimurium chromosome. The genes are categorized as follows (Kimbrough and Miller, 2002): i) export apparatus components; ii) needle complex structural components; iii) translocons; iv) regulators; v) effectors, and vi) chaperones. i) Export apparatus components. These are proteins that constitute the core export apparatus of virulence-associated secretion systems; since the majority of these proteins are either known or predicted to be integral membrane proteins, the core of this apparatus is believed to be located in a central pore within the base of the needle complex, where it facilitates the secretory-independent export of distal needle complex components and effector proteins; this group of proteins includes SpaO, P, Q, R, S, InvA, InvC, OrgB. All these genes are present in C. violaceum, except OrgB. ii) Needle complex structural components. Needle complex is the term given to the physical structure that can be isolated and visualized by transmission electron microscopy; purified needle complex is composed of at least the following protein components: PrgH, I, J, K, and InvG. All these genes are present in C. violaceum. iii) Translocons. The movement of effector proteins across the eukaryotic membrane is termed translocation; this process requires three proteins: SspB, C and D (also called SipBCD), which constitute a physiological structure, termed the translocon; the translocon proteins are believed to insert and form a pore in the eukaryotic cell membrane; in the absence of any one of these components, effector proteins are unable to cross the eukaryotic membrane and are secreted into the supernatant instead. All these genes (sipB, sipC, and sipD) are present in C. violaceum. iv) Regulators. Regulators of SPI1 gene expression function to restrict expression of the TTSS to specific locations within the host, and they coordinate the assembly process of the secretion apparatus; many of these regulators are encoded within SPI1 (InvF, HilA, HilD, SirC, SprB), while others, including PhoP/PhoQ and SirA/BarA, are encoded elsewhere in the chromosome. The genes InvF, HilA, and BarA are present in C. violaceum. v) Effectors. SPI1 TTSS translocates several effector proteins directly into the cytoplasm of eukaryotic cells, where they subvert cellular processes to favor bacterial colonization; while some of these are encoded within SPI1 (SspA/SipA, SptP, AvrA), many are encoded elsewhere in the chromosome (SopA, B, D, E, E2, SspH1, SlrP). vi) Chaperones. Chaperones are small, acidic, mostly alpha-helical proteins, which facilitate the efficient secretion and translocation of specific effector proteins; chaperones that are encoded on the island include SicA, InvB, and SicP. The genes SicA and InvB are present in C. violaceum. It has been shown that Fis, a DNA nucleoid-associated protein in Salmonella typhimurium, plays a pivotal role in the expression of HilA and InvF, two activators of SPI1 genes (Wilson et al., 2001). ORF CV37612 in C. violaceum codes for a protein with high similarity to Fis. Although C. violaceum has acquired most components of a S. typhimurium-like TTSS, some key genes, such as InvI, InvH and SicP, are absent. The lack of these genes may account for the generally poor ability of C. violaceum to infect humans. CONCLUDING REMARKS The abundance and diversity of transport-related membrane proteins encoded in the C. violaceum genome has allowed this bacterium to adapt to a wide range of environments in tropical and subtropical regions. Besides the possibility to provide insights on the many mechanisms and strategies used by microorganisms to interact and adapt to unfavorable and extreme environmental conditions, a more detailed analysis of the transport capabilities encoded in C. violaceum may provide a valuable source of genes with biotechnological potential, as exemplified by heavy metal transporters, which can be exploited in bioremediation. ACKNOWLEDGMENTS This study was supported by grants from the Ministério da Ciência e Tecnologia (MCT) and the Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq). T.B. Grangeiro had a research fellowship from CNPq (PQ-2B). REFERENCES Agranoff, D.D. and Krishna, S. (1998). Metal ion homeostasis and intracellular parasitism. Mol. Microbiol. 28: 403-412. Andrews, S.C., Smith, J.M.A., Guest, J.R. and Harrison, P.M. (1990). Genetic and structural characterization of the bacterioferritin of Escherichia coli. Biochem. Soc. Trans. 18: 658-659. Andrews, S.C., Findlay, J.B.C., Guest, J.R., Harrison, P.M., Keen, J.N. and Smith, J.M.A. (1991). Physical, chemical and immunological properties of the bacterioferritins of Escherichia coli, Pseudomonas aeruginosa and Azotobacter vinelandii. Biochim. Biophys. Acta 1078: 111-116. Andrews, S.C., Le Brun, N.E., Barynin, V., Thomson, A.J., Moore, G.R., Guest, J.R. and Harrison, P.M. (1995). Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 270: 23268-23274. Benz, R. and Bauer, K. (1988). Permeation of hydrophilic molecules through the outer membrane of gram-negative bacteria. Review on bacterial porins. Eur. J. Biochem. 176: 1-19. Campuzano, V., Montermini, L., Molto, M.D., Pianese, L., Cossee, M., Cavalcanti, F., Monros, E., Rodius, F., Duclos, F., Monticelli, A., Canizares, J., Koutnikova, H., Bidichandani, S.I., Gellera, C., Brice, A., Trouillas, P., Demichele, G., Filla, A., Defrutos, R., Palau, F., Didonato, S., Mandel, J.L., Cocozza, S., Koenig, M. and Pandolfo, M. (1996). Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271: 1423-1427. Carniel, E. (2001). The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3: 561-569. Cavadini, P., O’Neill, H.A., Benada, O. and Isaya, G. (2002). Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 11: 217-227. Cellier, M., Prive, G., Belouchi, A., Kwan, T., Rodrigues, V., Chia, W. and Gros, P. (1995). Nramp defines a family of membrane proteins. Proc. Natl. Acad. Sci. USA 91: 10089-10093. Crosa, J.H. (1997). Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61: 319-336. Durr, A., Cossee, M., Agid, Y., Campuzano, V., Mignard, C., Penet, C., Mandel, J.L., Brice, A. and Koenig, M. (1996). Clinical and genetic abnormalities in patients with Friedreich’s ataxia. New Engl. J. Med. 335: 1169-1175. Frolow, F., Kalb, A.J. and Yariv, J. (1994). Structure of a unique two-fold symmetric haem-binding site. Nat. Struct. Biol. 1: 453-460. Galan, J. and Collmer, A. (1999). Type III secretion machines; bacterial devices for protein delivery into host cells. Science 284: 1322-1328. Gibson, T.J., Koonin, E.V., Musco, G., Pastore, A. and Bork, P. (1996). Friedreich’s ataxia protein: phylogenetic evidence for mitochondrial dysfunction. Trends Neurosci. 19: 465-468. Govoni, G. and Gros, P. (1998). Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47: 277-284. Greenwood, K.T. and Luke, R.K. (1978). Enzymatic hydrolysis of enterochelin and its iron complex in Escherichia coli K-12. Properties of enterochelin esterase. Biochim. Biophys. Acta 525: 209-218. Hantash, F.M. and Earhart, C.F. (2000). Membrane association of the Escherichia coli enterobactin synthase proteins entB/G, entE, and entF. J. Bacteriol. 182: 1768-1773. Harrison, P.M. and Arosio, P. (1996). The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275: 161-203. Higgins, C.F. (2001). ABC transporters: physiology, structure and mechanism - an overview. Res. Microbiol. 152: 205-210. Hueck, C.J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62: 379-433. Janakiraman, A. and Slauch, J.M. (2000). The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35: 1146-1155. Jap, B.K. and Walian, P.J. (1990). Biophysics of the structure and function of porins. Q. Rev. Biophys. 23: 367-403. Jeanteur, D., Lakey, J.H. and Pattus, F. (1991). The bacterial porin superfamily: sequence alignment and structure prediction. Mol. Microbiol. 5: 2153-2164. Kimbrough, T.G. and Miller, S.I. (2002). Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4: 75-82. Komoriya, K., Shibano, N., Higano, T., Azuma, N., Yamaguchi, S. and Aizawa, S. (1999). Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34: 767-779. Koster, W. (2001). ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152: 291-301. Koutnikova, H., Campuzano, V., Foury, F., Dolle, P., Cazzalini, O. and Koenig, M. (1997). Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet. 16: 345-351. Le Brun, N.E., Andrews, S.C., Guest, J.R., Harrison, P.M., Moore, G.R. and Thomson, A.J. (1995). Identification of the ferroxidase centre of Escherichia coli bacterioferritin. Biochem. J. 312: 385-392. Madigan, M.T., Martinko, J.M. and Parker, J. (2002). Brock Biology of Microorganisms. Prentice Hall, Pearson Education, Inc., Upper Saddle River, NJ, USA. Mecsas, J. and Strauss, E.J. (1996). Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg. Infect. Dis. 2: 271-288. Nierman, W.C., Feldblyum, T.V., Laub, M.T., Paulsen, I.T., Nelson, K.E., Eisen, J.A., Heidelberg, J.F., Alley, M.R., Ohta, N., Maddock, J.R., Potocka, I., Nelson, W.C., Newton, A., Stephens, C., Phadke, N.D., Ely, B., DeBoy, R.T., Dodson, R.J., Durkin, A.S., Gwinn, M.L., Haft, D.H., Kolonay, J.F., Smit, J., Craven, M.B., Khouri, H., Shetty, J., Berry, K., Utterback, T., Tran, K., Wolf, A., Vamathevan, J., Ermolaeva, M., White, O., Salzberg, S.L., Venter, J.C., Shapiro, L., Fraser, C.M. and Eisen, J. (2001). Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98: 4136-4141. Pao, S.S., Paulsen, I.T. and Saier, M.H. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1-34. Putman, M., van Veen, H.W. and Konings, W.N. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64: 672-693. Sackin, H. (1995). Mechanosensitive channels. Annu. Rev. Physiol. 57: 333-353. Saier, M.H. (2000). A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64: 354-411. Usher, K.C., Ozkan, E., Gardner, K.H. and Deisenhofer, J. (2001). The plug domain of FepA, a TonB-dependent transport protein from Escherichia coli, binds its siderophore in the absence of the transmembrane barrel domain. Proc. Natl. Acad. Sci. USA 98: 10676-10681. Vasconcelos, A.T.R., Almeida, D.F., Hungria, M., Guimarães, C.T., Antônio, R.V., Almeida, F.C., Almeida, L.G.P., Almeida, R., Alves-Gomes, J.A., Andrade, E.M., Araripe, J., Araújo, M.F.F., Astolfi-Filho, S., Azevedo, V., Baptista, A.J., Bataus, L.A.M., Batista, J.S., Beló, A., van den Berg, C., Bogo, M., Bonatto, S., Bordignon, J., Brigido, M.M., Brito, C.A., Brocchi, M., Burity, H.A., Camargo, A.A., Cardoso, D.D.P., Carneiro, N.P., Carraro, D.M., Carvalho, C.M.B., Cascardo, J.C.M., Cavada, B.S., Chueire, L.M.O., Creczynski-Pasa, T.B., Cunha-Junior, N.C., Fagundes, N., Falcão, C.L., Fantinatti, F., Farias, I.P., Felipe, M.S.S., Ferrari, L.P., Ferro, J.A., Ferro, M.I.T., Franco, G.R., Freitas, N.S.A., Furlan, L.R., Gazzinelli, R.T., Gomes, E.A., Gonçalves, P.R., Grangeiro, T.B., Grattapaglia, D., Grisard, E.C., Hanna, E.S., Jardim, S.N., Laurino, J., Leoi, L.C.T., Lima, L.F.A., Loureiro, M.F., Lyra, M.C.C.P., Madeira, H.M.F., Manfio, G.P., Maranhão, A.Q., Martins, W.S., di Mauro, S.M.Z., Medeiros, S.R.B., Meissner, R.V., Moreira, M.A.M., Nascimento, F.F., Nicolás, M.F., Oliveira, J.G., Oliveira, S.C., Paixão, R.F.C., Parente, J.A., Pedrosa, F.O., Pena, S.D.J., Pereira, J.O., Pereira, M., Pinto, L.S.R.C., Pinto, L.S., Porto, J.I.R., Potrich, D.P., Ramalho-Neto, C.E., Reis, A.M.M., Rigo, L.U., Rondinelli, E., Santos, E.B.P., Santos, F.R., Schneider, M.P.C., Seuanez, H.N., Silva, A.M.R., Silva, A.L.C., Silva, D.W., Silva, R., Simões, I.C., Simon, D., Soares, C.M.A., Soares, R.B.A., Souza, E.M., Souza, K.R.L., Souza, R.C., Steffens, M.B.R., Steindel, M., Teixeira, S.R., Urmenyi, T., Vettore, A., Wassem, R., Zaha, A. and Simpson, A.J.G. (2003). The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 100: 11660-11665. Wilson, R.L., Libby, S.J., Freet, A.M., Boddicker, J.D., Fahlen, T.F. and Jones, B.D. (2001). Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39: 79-88. Zhou, D., Hardt, W. and Galan, J.E. (1999). Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Inf. Immun. 67: 1974-1981. |
|