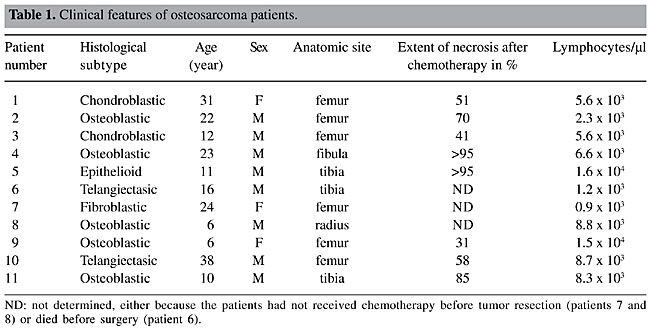
ABSTRACT. Osteosarcoma is the commonest type of primary malignant bone tumor, frequently found in adolescents at sites of rapid bone growth. Despite current management protocols, up to half of the patients succumb to this disease. Moreover, there is no well-characterized molecular marker for diagnosis and prognosis. Since phage display methodology allows the selection of human antibody fragments with potential use in clinical applications, we applied this procedure to construct a recombinant Fab (antigen binding fragment) library from patients with osteosarcoma. We used peripheral blood lymphocyte total RNA from 11 osteosarcoma patients and cloned recombinant Fab representing the µ, g and k chain antibody repertoires of these individuals. The resulting library was cloned in the pComb3X vector and attained 1.45 x 108 different functional forms. BstO I fingerprinting and DNA sequencing analysis of randomly selected clones revealed the diversity of the library, demonstrating that Fab harbors Vk chains from subgroups I to V, biased towards the A27 fragment, as normally reported for the human repertoire. Analysis of the VH repertoire revealed that our library has a slight bias towards the VH4 family, instead of the usually reported VH3. This is the first description of a phage display library from osteosarcoma patients. We believe these human Fab fragments will provide a valuable tool for the study of this neoplasia and could also contribute to improvements in the diagnosis of this disease. Key words: Phage display, Fab, Osteosarcoma, Recombinant antibody, Human antibody repertoire INTRODUCTION Antibodies have been used for antigen detection and therapeutics, and their specificity combined with low toxicity make them a promising pharmaceutical commodity (Berger et al., 2002). Currently, they comprise the second-largest category of biological medicines in clinical development, after vaccines (Chester et al., 2004). Successful examples include rituximab, approved by the FDA since 1997, an anti-CD20 antibody that is now an integral component of many treatment strategies for non-Hodgkin’s lymphoma (Seymour, 2004), and OKT3, an anti-CD3 that is widely used to reduce graft rejection (Cosimi et al., 1981). Therapeutic use of antibodies is limited by methodological constraints. They are usually obtained by immunization of experimental animals with target antigens; after screening, a specific antibody-producing hybridoma is identified (Kohler and Milstein, 1975). Although well established, this technology is laborious, and it is biased by the experimental model immune system, making it difficult to obtain a high-affinity antibody against conserved mammal proteins. Additionally, the heterologous proteins are often immunogenic for humans, preventing their therapeutic use (Maranhão and Brigido, 2000). Several examples of clinical antibody humanization to bypass this bottleneck have been described (Morrison and Oi, 1989); they minimize the human anti-murine antibody response by replacing murine sequences with human framework homologous sequences. The construction and selection of antibody combinatorial libraries displayed on filamentous phage surfaces became an alternative to this approach (McCafferty et al., 1990). In this technique, the repertoire of V genes of one or more individuals is amplified with primers covering all V gene families, giving rise to human antibodies. The library is generated by a random combination of variable light (VL) and variable heavy (VH) chain genes produced as antigen binding (Fab) or single chain variable (scFv) antibody fragments (Brigido and Maranhão, 2001). Theoretically, each clone codes for a specific antigen-binding site, corresponding to a natural repertoire, increased by an artificial domain combination that extrapolates the individual repertoire. This panning procedure mimics the B-cell clonal selection system in vitro by specifically enriching phage particles that display antibodies with a desired specificity (Barbas III et al., 1991). Several human antibody combinatorial libraries displayed on filamentous phage surface have been built by various groups, from either naive (Griffiths et al., 1993; de Haard et al., 1999; Lu et al., 2002) or immunized/infected repertoires (Portolano et al., 1993; Wu et al., 2001); this system has been applied to select antibodies against different antigens, including melanoma (Cai and Garen, 1995), colorectal (Somers et al., 2002) and prostate (Mintz et al., 2003) cancer proteins. Osteosarcoma is the most common primary malignant bone tumor, predominantly affecting children and adolescents; it is mainly found in areas with a high rate of skeletal growth. These tumors typically arise in the metaphyseal regions of long bones. The distal femur, proximal tibia and proximal humerus are the most common sites (Dahlin, 1975). Almost all osteosarcomas are high grade and have a poor prognosis. Usually, when the initial osteogenic sarcoma diagnosis occurs, tumors are large, and numerous lung micrometastases are already present (Jesus-Garcia Filho, 1992). Once metastases are diagnosed, the potential for cure is markedly decreased, and only 10% of the patients with this malignancy achieve a long-term disease-free interval (Meyers et al., 1993). In recent years, the immunocytochemical subclassification of osteosarcoma has significantly enhanced the accuracy of pathological diagnosis (Ueda et al., 1993); however, a specific marker has not been found and several studies have been made on the delineation of carcinogenesis molecular aspects. We developed a combinatorial Fab phage display library from randomly combined variable (V) region antibody genes of 11 osteosarcoma patients. This library could help us find new markers for diagnosis and even for the development of new treatment strategies. MATERIAL AND METHODS Patients We obtained peripheral blood lymphocytes (PBL) from 11 osteosarcoma patients attended at the Sarah Hospital for Rehabilitation in Brasília from 2000 to 2001. This study was previously approved by the hospital Ethics Committee. Patients’ features are shown in Table 1.
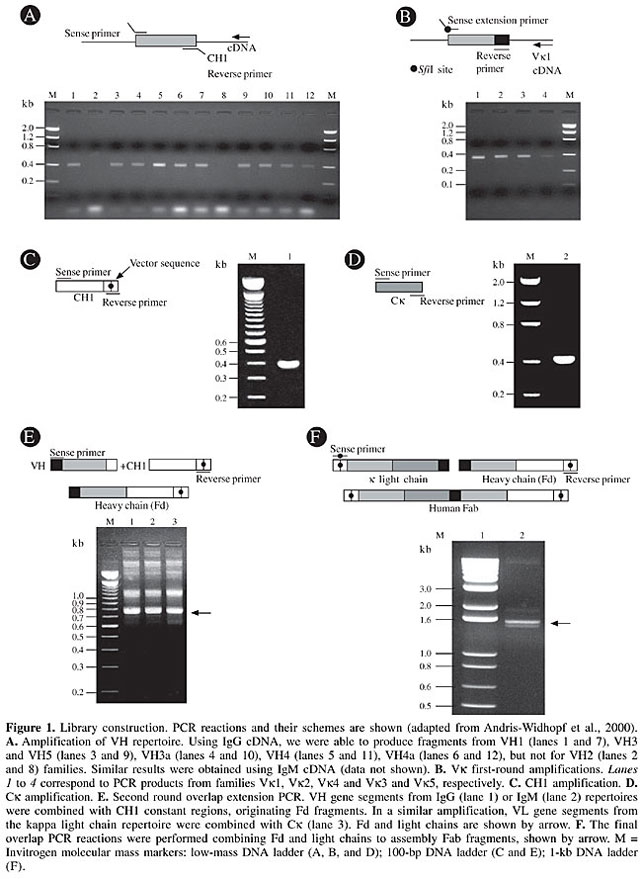
Library construction Total RNA from osteosarcoma patients was used to prepare a Fab combinatorial library displayed on the phage surface. The protocol involves four consecutive steps, as follows: i) synthesis of specific first-strand cDNAs; ii) synthesis by polymerase chain reaction (PCR) of VH genes for IgG and IgM classes and of the kappa light chain genes and their respective constant regions; iii) overlapping PCR to join each variable chain coding fragment with the respective constant region and a second PCR to obtain the complete Fab with the introduction of cloning sites at the extremities; iv) ligation of Fab into the pComb3X phagemid vector (Scott and Barbas III, 2000). A total of 1 x 107 lymphocytes from each patient was used to prepare the RNA using the QIAamp® RNA Blood Mini Kit (Qiagen). RNAs were pooled together and used to synthesize three different cDNAs (SuperScript System, Invitrogen), using specific primers to heavy and light chain immunoglobulins (Marks et al., 1991). Library construction was done as described by Andris-Widhopf et al. (2000). Briefly, we performed two sets of six amplifications, using sense primers for VH fragments, combined with gamma (g) or mi (µ) anti-sense primers. Similarly, VL gene fragments were obtained, using four sense primers, covering the whole kappa repertoire. These primers were used with a single 3’ oligonucleotide targeting kappa constant region (Ck). Primers used in this first round PCR were designed for the assembly of a heavy chain (Fd) fragment after a second round PCR. For this purpose, g and µ VH fragments were fused to a g constant region (CH1). The complete light chains were made by fusing VLs to a Ck fragment in another second round PCR. Fd fragments and kappa light chains were ultimately shuffled in a final overlap PCR. The final Fab coding fragments were digested with SfiI and cloned into the pComb3X, precipitated with ethanol, resuspended in 15 µl water and used to transform XL1-Blue E. coli (Stratagene) by electroporation, as described by Rader et al., 2000. After transformation, 5 ml SOC medium was added, and the culture was incubated for 1 h at 37°C at 250 rpm. At this point, culture aliquots were plated on LB agar/carbenicillin (50 µg/ml) to titer library size, which was calculated by counting the number of carbenicillin-resistant colonies per microgram DNA. SB medium (10 ml), containing carbenicillin (20 µg/ml) and tetracycline (10 µg/ml), was added to the transformed culture. After additional incubation under the same conditions, 100 ml SB medium, supplemented with carbenicillin (50 µg/ml) and tetracycline (10 µg/ml), was added. After 1 h of incubation, as above, 1012 plaque-forming units of VCSM13 helper phage (Stratagene) were added and the culture was shaken for 1 h at 37°C and 250 rpm. Kanamycin (70 µg/ml) was then added, and the culture was incubated overnight at 37°C and 250 rpm. Phages were obtained from the culture medium by polyethylene glycol/NaCl precipitation and were resuspended in TBS containing 1% BSA. Sequence analysis In order to prepare phagemid DNA, 96 different clones were cultured in a deep well plate containing 1 ml 2x YT medium with carbenicillin (100 µg/ml) and glucose (0.5% p/v) in each well. The cultures were incubated for 20 h at 37°C/320 rpm. Cells were recovered by centrifugation (3,000 g for 6 min), and the pellets were resuspended in 240 µl GET (0.92% glucose, 10 mM EDTA, pH 8.0, and 26 mM Tris-HCl) and harvested; an additional 80 µl GET buffer was then added. Sixty microliters of this suspension was transferred to a polypropylene plate (round bottom), treated with 2.5 µl RNAseA (10 mg/ml), and then 60 µl 1% 0.2 M NaOH SDS was added. Complete homogenization was obtained by inverting the sealed plate 30 times. To complete cell lysis, the plates were incubated at room temperature for 10 min. After this, 80 µl 3 M KOAc was added; the material was homogenized and incubated for an additional 10 min and then centrifuged at 3000 g for 8 min. The supernatants were transferred to a new 96-well V-bottom plate and filtered through a Millipore membrane plate by centrifuging at 3000 g for 5 min. The DNA was precipitated by adding isopropanol (100 µl), followed by centrifugation at 3000 g for 45 min. The pellet was washed with 70% ethanol, dried and resuspended in 30 µl milliQ water. Library quality was assessed by digestion of carbenicillin resistant clones DNA with SfiI, and its diversity was evaluated by BstO I restriction pattern and DNA sequencing. The sequencing reactions were prepared with specific reverse primers CH1 (5’ CGCCTGAGTTCCACGACACC 3’) or MMB5 (5’ CGTTTGCCATCTTTTCATAATC 3’) for VH and primers Ck (5’ AGAGGAGTCCAGATTTCA 3’) or MMB4 (5’ GCTTCCGGCTCGTATGTTGTGT 3’) for Vk genes, using the ET Terminator kit (Amershan-Pharmacia), following manufacturers’ instructions. Sequencing reactions were resolved in a MegaBACE 1000 automatic sequencer (Molecular Dynamics). V gene sequence determinations were based on Phred basecalling (Ewing and Green, 1998) and chromatograms were used for manual checking of sequence ambiguities. Sequence alignments and translations were made with the program BioEdit (Hall, 1999). V gene families were assigned using the Ig-Blast server at NCBI (http://www.ncbi.nlm.nih.gov). The Kabat numbering and CDR definition were adopted from Andrew’s web site (www.bioinf.org.uk/abs/). RESULTS Human Fab phage display library construction Eleven patients with a confirmed diagnosis of osteosarcoma, attended at the Sarah Hospitals for Rehabilitation from September 2000 to April 2001, were the immunoglobulin gene donors. All of them had conventional high-grade intramedular osteosarcoma, with various histological features (Table 1). The different behaviors after chemotherapy treatment may reflect their individual oncogenic characteristics. This may be attributed to very diverse immunological responses to the tumors, which contribute to the generation of a very diverse Fab library. In fact, the pathological and clinical heterogeneity of osteosarcoma has long been recognized by those regularly involved in the management of these tumors (Whelan, 1997). The Fab heavy chain (Fd) and k light chain genes were obtained using cDNA synthesized from the total RNA of PBL of the 11 osteosarcoma patients. Twelve separate VH and four Vk PCR reactions were performed to optimize the amplifications of V gene rearrangements. The resulting PCR products, corresponding to VH (Figure 1A) and Vk (Figure 1B) gene fragments, account for a large fraction of the rearranged variable region repertoire of these patients. All the primary products of PCR amplification of the variable and constant regions (Figure 1C for CH1 and 1D for Ck) ranged from 350 to 400 bp in size. In the overlapping second round of PCR, the resulting variable genes were pooled and joined together, along with the constant region, generating PCR products ranging from 750 to 800 bp (Figure 1E). The third PCR round consisted of a final overlap amplification of the complete human dicistronic Fab gene, generating fragments of 1.5 to 1.6 kb (Figure 1F). The generated Fabs were cloned into the pComb3X phage display vector, after digestion with the restriction enzyme SfiI. Since the PCR primers incorporated two asymmetric SfiI sites, this strategy produced a directional ligation of the SfiI digested Fab, in frame with ompA and pelB leader sequences, to direct expression of antibody light and heavy chain-pIII fusion proteins, under the control of the lac promoter. The gene III Fd fusions included His and influenza hemagglutinin epitope tags. In addition, an amber stop codon, TAG, was inserted after the hemagglutinin tag, allowing the production of soluble Fab in a nonsuppressor E. coli strain (Scott and Barbas III, 2000). OmpA and pelB signal sequences directed the Fab to periplasmatic cell compartment and the resulting Fab-coat protein fusion was incorporated onto the surface of M13 Fab-phage fusion particles after helper phage infection. The resulting combinatorial library reached 1.8 x 108 Fabs, forming potential paratopes able to recognize an equivalent number of epitopes.
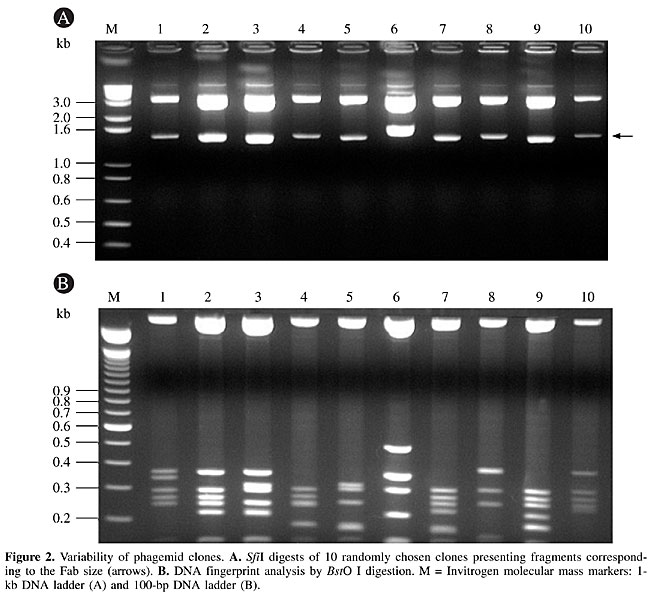
Library diversity V gene usage To assess the quality and diversity of the library, 10 randomly selected Fab clones were digested with SfiI and BstO I restriction enzymes. The SfiI digestion yielded 1.5- to 1.6-kb fragments, corresponding to the expected Fab insert. The BstO I fingerprint profiles confirmed the diversity of the library (Figure 2). All the analyzed clones presented the correct Fab size, and all of them had distinct BstO I restriction fragment patterns. Since all clones had the same CH1 and Ck region genes and vector backbones, differences in the restriction patterns reflected differences in the VH or Vk rearranged genes. Thus, the library shows diversity at the V gene level (Figure 2).
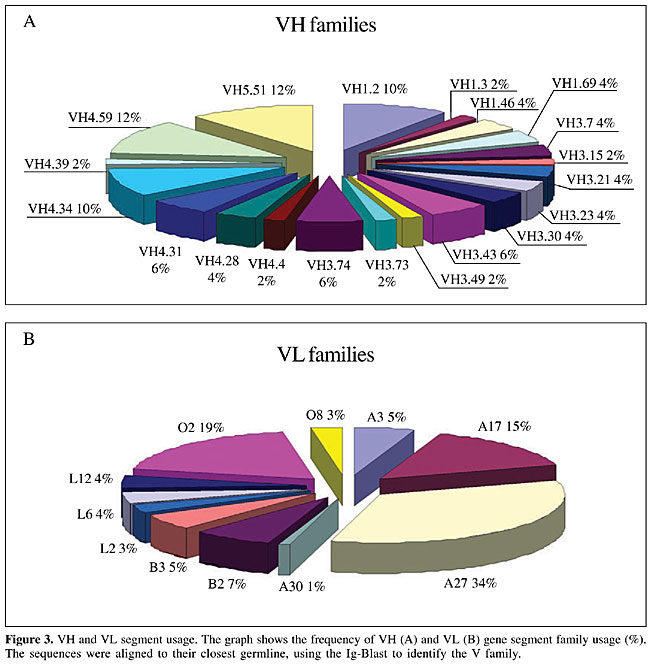
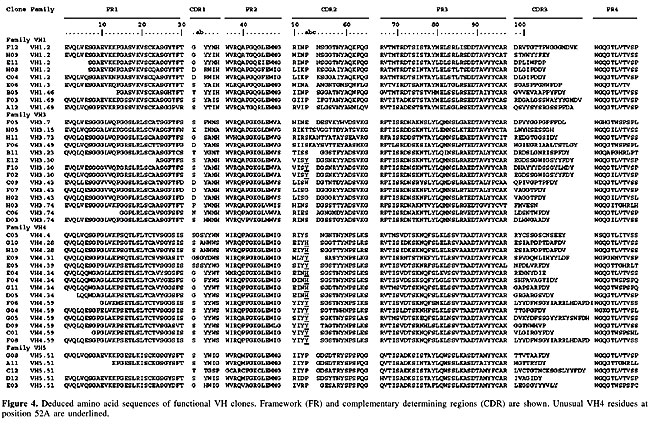
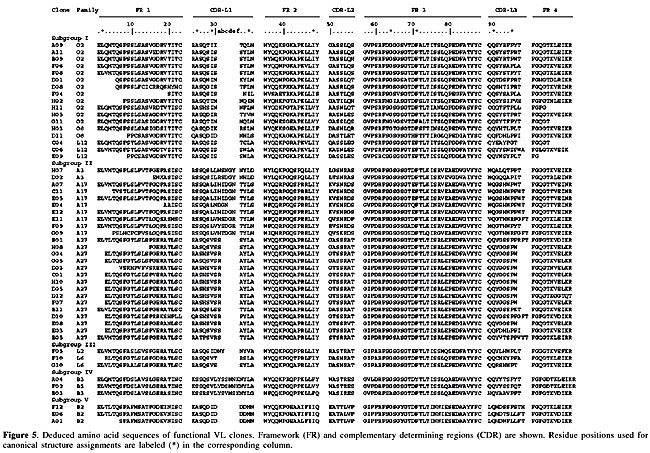
A further investigation included the sequencing of several V gene clones. Sequence comparison was made using the Ig Blast defined patients’ V gene family repertoire usage. We sequenced either VH or Vk in 125 V regions (51 VH and 74 Vk). Although all the clones in our library bear a full length insert, 80.8% of them had a functional VH/VL fusion Fab, leading to a functional library size of 1.45 x 108. The VH repertoire, evaluated among 51 randomly chosen clones, showed 20 different VH subfamilies (Figure 3A). No significant bias was observed, but there was a slight predominance of clones using genes from the VH4 family (35.4%), which differs from the reported predominance of the VH3 family (33.3% of our clones) in the normal human repertoire (Cook and Tomlinson, 1995). No VH2 amplification was observed. The failure in detecting VH2 members, notwithstanding the use of specific primers, can be explained by the low frequency of usage of this family (Knappik et al., 2000), though polymorphism or even primer design defects cannot be discarded. Concerning Vk genes (Figure 3B), among 74 sequenced clones, 11 different families were found, with 33% predominance of A27, similar to what was found in a previous report (Tomlinson et al., 1995). In our sequences, the second most frequent family was O2 (18.9%), followed by A17 (14.8%) (Figure 3B). The resulting library contained VH genes from four different families (VH families 1, 3, 4, and 5), and k light chain genes from subgroups I, II, III, IV, and V. Complementary determining (CDR) and framework region assignments were based on Kabat definition. Only high quality sequences were used for translation (Figures 4 and 5). The analysis was successfully accomplished in 43 VH and 51 Vk sequences, respectively.
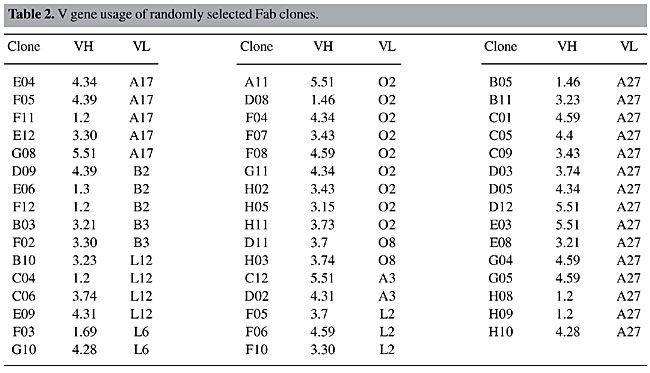
Random Fab assembly was evident in the analysis of VH and VL pairs of 47 individual clones (Table 2). Most of the clones were assembled with different variable genes; only three pairs of clones (D12 and E03, G04 and G05 and H8 and H9) shared the same families of both VH and VL genes even though they accumulated other differences in framework or CDR sequences. Three pairs of clones presented the same VH sequence, H08-C04, F07-H02 and G10-H10, but they had different VL genes (Figure 4 and Table 2).
VH diversity Heavy chain gene analysis revealed a great variability of family usage (Table 2, Figure 3). Most of the clones presented the canonical residues at clue positions. One of the exceptions is clone E04, which had a serine instead of a glycine at position 26 (G26S). Position 27 is expected to show F, Y, S, D, G, or T. Some of our clones presented different residues: H (clone B05), A (clones E09 and D09) and E (clone F04). We also found differences in residue 94: T, S, K, A, and L, instead of the canonical R, G or N (Chothia et al., 1989). The heterogeneity of CDRH3 was remarkable. Its length varied from 6 to 20 amino acid residues. In our clones, the most frequent CDR sizes were 8 (23%), 9, 11, and 13 (11.6% each) residue lengths. Classifying canonical loop 1 structures, our clones were mainly type 1 (90%), followed by some with type 2 (5%) and 3 (5%) loops. These frequencies are similar to those known for human antibody canonical H1 conformations (Almagro et al., 1997). Assignment of H2 canonical conformation was not possible for nearly half of the sequenced clones (Figure 4). The recurrence of histidine and tyrosine residues at position 52a has not been previously reported, but we found it to be frequent. This observation may reflect a polymorphism of the Brazilian population or a donor patient repertoire bias. It is noteworthy that these non-usual residues are found in the VH4-derived Fabs, which were 48% of our clones. The H2 loops had type 1 (30%), 2 (12%), 3 (9%), or 4 (7%) canonical conformations. VL diversity Key residues at positions 2, 25, 29, 33, and 71 contribute to the L1 structure. The 51 sequences listed in Figure 4 are according to these key presentations or have previously described mutations (three clones from the O2 family had changes at position 33: clone H05: L33V, and clones F04 and H02: L33I, and one clone, A04, with F71Y). All functional germ line Vk segments had a single canonical L2 structure, which contained an isoleucine at position 48, a glycine at site 64 and one residue between residues 50 and 52. Among our sequences, we observed five clones with rare changes at this position. Clones G11 and H11 (from the VLO2 family) and clone F02, belonging to VLB3, had a conservative change from isoleucine to valine (I48V) and both A04 and B03 clones (VLB3) presented a non-conservative substitution from isoleucine to phenylalanine (I48F), which is reported to occur in less than 1% of the Vk sequences (Tomlinson et al., 1995). The B03 clone also presented a conservative change from glycine to alanine (G64A) at position 64, while clone B11 had an aspartic acid (G64D) at this position. The most variable region was CDRL3, due to V-J gene segment rearrangements. The joining process involved trimming and repair of the V and J segments, and there was also N addition. CDR3 can vary in length from 1 to 11 residues, with most of the sequences presenting between 6 and 8 amino acid residues (Knappik et al., 2000). Among our patients, there was a predominance of CDRL3s with 9 (44.5%), followed by 8 (24.0%), 10 (20.4%) and 11 (11.1%) residues. Tomlinson and co-workers (1995) studied 634 VL genes and reported 67% with 6 residues, 19% with 7 and only 0.15% of the CDRL3s with 9 residues. In a study of 382 VK genes, Knappik and co-workers (2000) found 72.3% of all CDR3s with 8 amino acid residues, 7.3% with 7, 17.3% with 9, 1.3% with 10, and 1.8% with less than 7 residues in the CDRL3. There are five canonical structures for the L3, which vary according to the number of residues. Most of them have a glutamine residue at position 90 and a proline at position 95. Some of our clones had differences at those positions. One clone had a tyrosine at position 90 (Q90Y) and a proline at position 94 (C04 clone from the L12 family). The presence of a proline at positions 94 has been described in very few sequences (Chothia et al., 1989). Three of our clones had a proline at position 95A: C06, E11, G09. Three clones from the B2 family (D09, E06 and A01) had a leucine at this position (P95L) and one clone (B10, from the L12 family) had a P95F. DISCUSSION We cloned genes coding for the human antibody repertoire of osteosarcoma patients, for future use for the isolation of immunoglobulins against these tumor antigens. We attempted to maximize the library diversity by using primers optimized for VH (IgG and IgM) and Vk gene family mRNAs. Osteosarcoma antibody variable gene repertoires were cloned into a phage display vector, allowing combinatorial Fab assembly. Library size was compatible to the reported size for naive and immunized human phage displayed libraries, which range from 106 to 109 (Vaughan et al., 1996; Itoh et al., 2003). It has long been recognized that actual library size must correspond to the functional antibody forms. Thus, the real size is smaller than that found by counting recombinant colonies, since frameshifts, stop codons, or deletions can be generated by PCR or can be a product of non-productive immunoglobulin rearrangements. Our real library is formed of 81% functional clones, originating around 108 Fabs. This order of magnitude should permit successful antigen-binding antibody selection. Moreover, due to its origin, it is expected that this library is tumor specific, along with environmental and self-antigen-reacting antibodies. Even new forms, resulting from in vitro shuffling of VH and VL, enrich the library’s natural repertoire. Analysis of VH/VL pairs also revealed that none of these 47 randomly selected clones shared the same VH and VL sequences, being unique and hypothetically accounting for different specificities. Thus, the library size and its variability will certainly allow for the selection of useful anti-osteosarcoma antibodies and also immunoglobulins that recognize a great variety of epitopes, since non-immune libraries are being used to isolate antibodies against different antigens (de Haard et al., 1999). Our library will be used to select antibodies against bone tumor cell total protein preparations and also against their cell surface, targeting molecular markers. Ours is the first report of an osteosarcoma-patient antibody repertoire-derived library. The considerable variability of our library means that it should be useful for isolating Fabs capable of recognizing osteosarcoma antigens. These new molecules are potentially valuable tools in the detection of these tumor malignant cells and could be helpful for improved diagnosis and treatment. ACKNOWLEDGMENTS We are grateful to Isabella Simões and Fabricio Arraes for sequencing, Dr. Marcio Poças-Fonseca for English revision, Cid Alexandre Pereira for figure preparation, and the Sarah Network of Hospitals for Rehabilitation for financial support. REFERENCES Almagro, J.C., Hernandez, I., Ramirez, M.D.C. and Vargas-Madrazo, E. (1997). The difference between the structural repertoire of VH germline gene segments of mice and human: implication for the molecular mechanism of the immune response. Mol. Immunol. 34: 1199-1214. Andris-Widhopf, J., Steinberger, P., Fuller, R., Rader, C. and Barbas III, C.F. (2000). Generation of antibody libraries: PCR amplification and assembly of light- and heavy-chain coding sequences In: Phage Display Laboratory Manual (Barbas III, C.F., Burton, D.R., Scott, J.K. and Silverman, G.J., eds.). 1st edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA, pp. 9.1-9.22. Barbas III, C.F., Kang, A.S., Lerner, R.A. and Benkovic, S.J. (1991). Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 88: 7978-7982. Berger, M., Shankar, V. and Abbas, V. (2002). Therapeutic applications of monoclonal antibodies. Am. J. Med. Sci. 324: 14-30. Brigido, M.M. and Maranhão, A.Q. (2001). Bibliotecas apresentadas em fagos. Biotecnologia - Ciência e Desenvolvimento 26: 44-51. Cai, X. and Garen, A. (1995). Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: Selection of specific antibodies from single-chain Fv fusion phage libraries. Proc. Natl. Acad. Sci. USA 92: 6537-6541. Chester, K., Pedley, B., Tolner, B., Violet, J., Mayer, A., Sharma, S., Boxer, G., Green, A., Nagl, S. and Begent, R. (2004). Engineering antibodies for clinical applications in cancer. Tumor Biol. 25: 91-98. Chothia, C., Lesk, A.M., Tramontano, A., Levitt, M., Smith-Gill, S.J., Air, G., Sheriff, S., Padlan, E.A., Davies, D., Tulip, W.R., Colman, P.M., Spinelli, S., Alzari, P.M. and Poljak, R.J. (1989). Conformations of immunoglobulin hypervariable regions. Nature 342: 877-883. CooK, G.P. and Tomlinson, I.M. (1995). The human immunoglobulin VH repertoire. Immunol. Today 16: 237-242. Cosimi, A., Burton, R., Colvin, R., Goldstein, G., Delmonico, F., LaQuaglia, M., Tolkoff-Rubin, N., Rubin, R., Herrin, J. and Russell, P. (1981). Treatment of acute renal allograft rejection with OKT3 monoclonal antibody. Transplantation 32: 535-539. Dahlin, D.C. (1975). Pathology of osteosarcoma. Clin. Orthop. 111: 23-32. de Haard, H.J., van Neer, N., Reurs, A., Hufton, S.E., Roovers, R.C., Henderikx, P., Bruïne, A.P., Arends, J.W. and Hoogenboom, H. (1999). A large non-immunized human Fab fragment phage display that permits rapid isolation and kinetics analysis of high affinity antibodies. J. Biol. Chem. 274: 18218-18230. Ewing, B. and Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. Griffiths, A.D., Malmqvist, M., Marks, J.D., Bye, J.M., Embleton, M.J., McCafferty, J., Baier, M., Holliger, K.P., Gorick, B.D., Hughes-Jones, N.C., Hoogenboom, H.R. and Winter, G. (1993). Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 12: 725-734. Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis. Nucleic Acids Symp. 41: 95-98. Itoh, K., Inoue, K., Tezuka, T., Tada, H., Hashimoto, Y., Masuko, T. and Suzuki, T. (2003). Molecular structural and functional characterization of tumor suppressive anti-erbB-2 monoclonal antibody by phage display system. J. Biochem. 133: 239-245. Jesus-Garcia Filho, R. (1992). Tumores produtores de tecido ósseo. In: Manual de Tumores Músculo-Esqueléticos (Jesus-Garcia Filho, R. and Nery, C.A.S., eds.). Escola Paulista de Medicina, São Paulo, SP, Brasil, pp. 18-27. Knappik, A., Ge, L., Honegger, A., Pack, P., Fischer, M., Wellnhofer, G., Hoess, A., Wölle, J., Plünckthun, A. and Virnekas, B. (2000). Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CRDs randomized with trinucleotides. J. Mol. Biol. 296: 57-86. Kohler, G. and Milstein, C. (1975). Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256: 495-497. Lu, D., Jimenez, X., Zhang, H., Boleen, P., Witte, L. and Zhu, Z. (2002). Selection of high affinity human neutralizing antibodies to VEGFR2 from a large antibody phage display for antiangiogenesis therapy. Int. J. Cancer 97: 393-399. Maranhão, A.Q. and Brigido, M.M. (2000). Anticorpos humanizados. Biotecnologia - Ciência e Desenvolvimento 23: 38-43. Marks, J.D., Hoogenboom, H.R., Bonnert, T.P., McCafferty, J., Griffiths, A.D. and Winter, G. (1991). By-passing immunization of human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 222: 581-597. McCafferty, J., Griffiths, A.D., Winter, G. and Chiswell, D.J. (1990). Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348: 552-554. Meyers, P.A., Heller, G., Huvos, A., Applewhite, A., Sun, M. and LaQuaglia, M. (1993). Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J. Clin. Oncol. 11: 449-453. Mintz, P.J., Kim, J., Do, K.-A., Wang, X., Zinner, R.G., Cristofanilli, M., Arap, M.A., Hong, W.K., Troncoso, P., Logothetis, C.J., Pasqualini, R. and Arap, W. (2003). Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat. Biotechnol. 21: 57-63. Morrison, S.L. and Oi, V.T. (1989). Genetically engineered antibody molecules. Adv. Immunol. 44: 65-92. Portolano, S., McLachlan, S.M. and Rapoport, B. (1993). High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J. Immunol. 151: 2839-2851. Rader, C., Steinberger, P. and Barbas III, C.F. (2000). Selection from antibody libraries In: Phage Display Laboratory Manual (Barbas III, C.F., Burton, D.R., Scott, J.K. and Silverman, G.J., eds.). 1st edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA, pp. 10.2-10.20. Scott, J.K. and Barbas III, C.F. (2000). Phage display vectors. In: Phage Display Laboratory Manual (Barbas III, C.F., Burton, D.R., Scott, J.K. and Silverman, G.J., eds.). 1st edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA, pp. 2.1-2.19. Seymour, J.F. (2004). New treatment approaches to indolent non-Hodgkin’s lymphoma. Semin. Oncol. 31: 27-32. Somers, V.A., Brandwijk, R.J., Joosten, B., Moerkerk, P.T., Arends, J.W., Menheere, P., Pieterse, W.O., Claessen, A., Scheper, R.J., Hoogenboom, H.R. and Hufton, S.E. (2002). A panel of candidate tumor antigens in colorectal cancer revealed by serological selection of a phage displayed cDNA expression library. J. Immunol. 169: 2772-2780. Tomlinson, I.M., Cox, J.P.L., Gherardi, E., Lesk, A.M. and Chothia, C. (1995). The structural repertoire of the human Vk domain. EMBO J. 14: 4628-4638. Ueda, Y., Roessner, A. and Grundmann, E. (1993). Pathological diagnosis of osteosarcoma: the validity of the subclassification and some new diagnostic approaches using immunohistochemistry. Cancer Treat. Res. 62: 109-124. Vaughan, T.J., Williams, AJ., Pritchard, K., Osbourn, J.K., Pope, A.R., Earnshaw, J.C., McCafferty, J., Hodits, R.A., Wilton, J. and Johnson, K.S. (1996). Human antibodies with sub-nanomolar affinities isolated from a large non-immunized Phage Display Library. Nat. Biotechnol. 14: 309-314. Whelan, J.S. (1997). Osteosarcoma. Eur. J. Cancer 33: 1611-1619. Wu, B.P., Xiao, B., Wan, T.M., Zhang, Y.L., Zhang, Z.S., Zhou, D.Y., Lai, Z.S. and Gao, C.F. (2001). Construction and selection of the natural immune Fab antibody phage display library from patients with colorectal cancer. World J. Gastroenterol. 7: 811-815. |
|