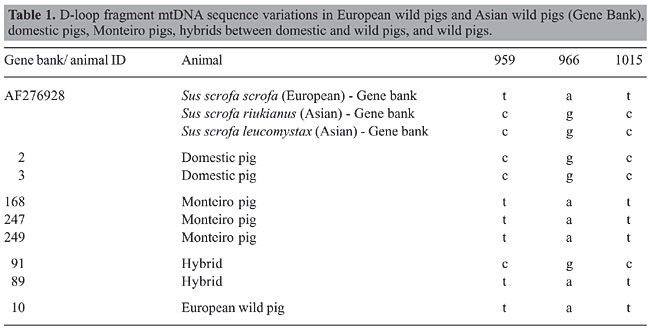
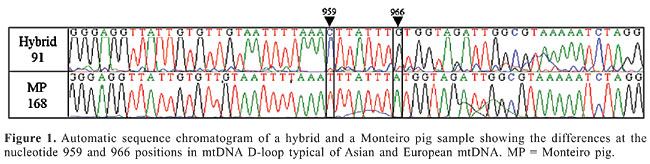
ABSTRACT. We examined the variation in mitochondrial DNA by sequencing the D-loop region in wild and domestic (large-white breed) pigs, in hybrids between domestic and wild pigs, and in Monteiro pigs. A D-loop fragment of approximately 330 bp was amplified by PCR. Sequencing of DNA amplicons identified haplotypes previously described as European and Asian types. Monteiro pigs and wild pigs had European haplotypes and domestic pigs had both European and Asian haplotypes. Key words: Exotic animals, Genetic diversity, Evolution, Molecular genetic INTRODUCTION The wild pig (Sus scrofa scrofa) is an introduced species in Brazil, imported from Uruguay and Argentina 20 years ago and more recently (1997) from France and Canada. Due to the large demand for exotic meat, it is currently crossed commercially with domestic pigs to produce hybrids (Lui, 2000). Pigs were domesticated about 10,000 years ago and wild and domestic pigs belong to the same species (Sus scrofa). Wild pigs are classified in the subspecies scrofa L. and the domestic swine in subspecies domestica L. Domestic swine normally present 2n = 38 and the wild pig has 2n = 36 (Bosma, 1976). Despite these differences in chromosome number, these animals can mate and produce fertile hybrids. The hybrid has 37 chromosomes, and it is morphologically similar to the wild pig (Lui, 2000). Hybrids can successfully breed with the wild and domestic pigs or with other hybrids, generating offspring with 36, 37 or 38 chromosomes. Brazil also has a local “wild” Sus scrofa breed, called the “Monteiro” pig, which descended from domestic stock brought to Brazil during the initial period of colonization (Silva and Mauro, 2002). Researchers have been studying and analyzing the genetic intraspecific and species-specific diversity of animals of economic interest and of wild animals. For studies of genetic differentiation, mitochondrial DNA (mtDNA) has been widely used as a tool to characterize races and determine genetic relationships (Brown et al., 1989; Pegoraro et al., 1996; Watanobe et al., 1999). We examined the variation in mtDNA of wild pigs, domestic swine, hybrids (wild x domestic), and “Monteiro” pigs, specifically looking at nucleotide polymorphism in the D-loop region. MATERIAL AND METHODS Blood was collected from the jugular vein into EDTA-containing vacuum tubes from a wild pig (enumerated 10 W), two hybrid pigs (identified as 89 and 91), two domestic pigs, offspring of a cross between a Large White female and a Duroc male, identified as 2D and 3D, and three “Monteiro” pigs (identified as MP168, MP247 and MP249). These samples were homogenized immediately. DNA was extracted and purified by applying the phenol-chloroform method (Sambrook et al., 1989). PCR of the D-loop region A fragment of the D-loop region was amplified by polymerase chain reaction (PCR) using the primers: MITL4 5’ - CCAAAAACAAAGCAGAGTGTAC - 3’ described by Okumura et al. (1996) and MITH4 5’ - AGGCATTTTCAGTGCCTTG - 3’, designed from the sequence deposited in Gen bank (AF276928 - www.ncbi.nlm.nih.gov). The PCR reaction was made in a final volume of 10 µL with the following: 3.0 mM MgCl2, one unit of Taq DNA polymerase, 0.2 mM of each dNTP, PCR buffer 10X (200 mM Tris-HCl, pH 8.4, 500 mM KCl), and 5 pmol of each primer. Amplification cycles consisted of 34 cycles each, consisting of 94°C for 30 s, 60°C for 45 s and 72°C for 60 s. DNA fragment amplification was confirmed by electrophoresis on 1% agarose gels stained with ethidium bromate and visualized under UV light. Sequencing of the D-loop region The purification of PCR products was accomplished using the QiaexII Gel Extraction Kit (QIAGEN Inc.). The fragment was sequenced using the “Big Dye” kit (Applied Biosystems) in an automatic ABI 377 sequencer (Molecular Dynamics) and was analyzed using Bioedit software, version 7.0.0 (Ibis Therapeutics). RESULTS AND DISCUSSION Figure 1 shows part of a chromatogram of a hybrid and a “Monteiro” pig mtDNA sample. Eight sequences were analyzed and compared with Sus scrofa mtDNA sequences previously deposited in Gen Bank. The results are shown in Table 1. We examined the D-loop fragment sequence of five S. scrofa domestica (2 domestic pigs and 3 Monteiro pigs) one S. scrofa scrofa and two S. scrofa scrofa x S. scrofa domestica. A total of 225 bases were sequenced from position 741 to 1091 of the mtDNA in all samples. Two haplotypes were identified with variation at three positions. The remainder of the sequence in all samples was identical to the sequence retrieved from Gen Bank. Polymorphic sequence variation was observed at the following positions: either t or c at 959, either a or g at 966, and either t or c at 1015. No heteroplasmic animals were observed. The patterns of D-loop sequences corresponded to previously reported alleles considered to represent European Sus scrofa domestica and Asian Sus scrofa riukyanus and Sus scrofa leucomystax origins (Table 1). Both alleles were found, but they were clearly segregated. Domestic swine exhibited the Asian haplotype, but all Monteiro pig samples exhibited the European haplotype. Asian breeds are known to be very efficient in their reproduction, having and rearing large litters. This has stimulated both research as well as the utilization of Asiatic pigs for genetic improvement of domestic swine. It has been reported that Large Whites carry both the European (Large White I) and Asian haplotypes (Large White II: Watanobe, 1999; Chen and Leibenguth, 1995). This is consistent with the results that we obtained for domestic swine. The European haplotypes of the three Monteiro pigs are also consistent with the history of the Monteiro pig in South America. These animals were brought from Europe during colonization and escaped from captivity. We suggest that the Monteiro pig is a descendent of early European stocks. The Sus scrofa scrofa individual had an mtDNA pattern typical for a European haplotype, as expected.
The hybrid animals were descendents of a wild-pig boar mated with a domestic female; they had a 37-chromosome genotype. The two hybrid animals had both patterns of mtDNA, suggesting a maternal ancestor of Asian origin in one animal and European in the other. Considering that the number of chromosomes is not always informative of the hybrid or pure origin of the pigs, the presence of Asiatic mtDNA can help to identify individuals descended from hybrids. Although we analyzed few animals in this study, domestic and hybrid pigs were demonstrated to originate from two different stocks (genetic sources). Our data indicate an important contribution of Asian pigs to the domestic pigs. Analyzing the number of female lineages carrying an Asian haplotype would help to understand the contribution of Asiatic genes to commercial pig herds in Brazil. We conclude that in the time elapsed since pigs were first domesticated, there have been no subspecies specific changes in the genotype/sequence of the mtDNA D-loop fragment that we characterized. Although Sus sus domesticus has been subject to genetic manipulation (selection), focused on reproductive and production traits, the sequence of this fragment of the D-loop region has not changed in a substantial way. ACKNOWLEDGMENTS We thank Dr. Alan Conley, University of Davis, California, for careful reading of the paper; Prof. Dr. Luiz Lehman Coutinho and the team of his laboratory, at ESALQ, where sequencing was carried out; Prof. Dr. José Maurício Barbante Duarte, from UNESP, Jaboticabal, for providing the Monteiro pig samples, and Paulo A. Tosta and João A. Boer for the excellent technical assistance. S.F. Grossi was the recipient of a doctoral fellowship from CNPq. REFERENCES Bosma AA (1976). Chromosomal polymorphism and G-banding patterns in the wild boar (Sus scrofa L.) from the Netherlands. Genetica 46: 391-399. Brown DR, Koehler CM, Lindberg GL, Freeman AE, et al. (1989). Molecular analysis of cytoplasmic genetic variation in Holstein cows. J. Anim. Sci. 67: 1926-1932. Chen H and Leibenguth F (1995). Restriction patterns of mitochondrial DNA in European wild boar and German Landrace. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 110: 725-728. Gen bank. (http://www.ncbi.nlm.nih.gov). Lui JF (2000). Estudo citogenético de javalis (Sus scrofa scrofa) e híbridos nas regiões sudeste e sul do Brasil. Rev. Educ. Contin. CRMV-SP 3: 43-48. Okumura N, Ishiguro N, Nakano M, Hirai K, et al. (1996). Geografic population structure and sequence divergence in the mitochondrial DNA control region of the Japanese wild boar (Sus scrofa leucomystax), with reference to those of domestic pigs. Biochem. Genet. 34: 179-189. Pegoraro L, Yang Z, Samake S, Meirelles FV, et al. (1996). Sequence comparison of mitochondrial tRNA genes and origin of light strand replication in Bos taurus and Nellore (Bos indicus) breeds. Anim. Genet. 27: 91-94. Sambrook J, Frietsch EF and Maniatis T (1989). Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA. Silva MP and Mauro R (2002). Utilización de pasturas nativas por mamíferos e herbívoros en el pantanal. Arch. Zootec. 51: 161-173. Watanobe T, Okumura N, Ishiguro N, Nakano M, et al. (1999). Genetic relationship and distribution of the Japanese wild boar (Sus scrofa leucomystax) and the Ryukyu wild boar (Sus scrofa riukiuanus) analysed by mitochondrial DNA. Mol. Ecol. 8: 1509-1512. |
|