| DNA extraction from fixed cytogenetic cell suspensions
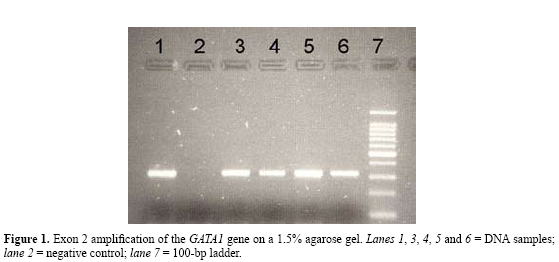
M.R. Amorim1, F.R. Vargas2, J.C. Llerena Junior3 and M.S. Pombo-de-Oliveira1 Genet. Mol. Res. 6 (3): 500-503 (2007) ABSTRACT. We developed a procedure for DNA extraction from small volumes of fixed cell suspensions previously prepared for conventional cytogenetic analysis. Good quality DNA was isolated with a fast and simple protocol using DNAzol reagent. This provided suitable DNA for various types of molecular analyses, including polymerase chain reaction, restriction fragment length polymorphism, denaturing high-performance liquid chromatography, and direct sequencing. This technique provides sufficient material for such test, which are important for diagnosis of neoplastic diseases in pediatric patients. Key words: DNA extraction, DNAzol, Cell suspension, Cytogenetic INTRODUCTION Ordinarily, cells are subjected to hypotonic treatment in cytogenetic preparations. A fixative solution (3:1 methanol:glacial acetic acid) is added to the cell suspension; usually the fixative removes lipids and denatures proteins, making the cell membrane remnants very fragile, which allows subsequent chromosome spreading and consequently exposes the DNA. These preparations are usually stored for years after chromosome analysis (Coleman and Tsongalis, 1997). We developed a protocol for DNA extraction from fixed human cell suspensions previously prepared for cytogenetic analysis. As we have been backtracking GATA1 mutations in newborns with Down syndrome, we tested whether frozen cell suspensions obtained for conventional cytogenetic analysis performed at birth would be suitable for molecular analyses. MATERIAL AND METHODS Samples Peripheral blood and/or bone marrow cell suspensions were obtained from 30 children with Down syndrome. They were previously submitted to a karyotype analysis from 2003 to 2006 at the Genetic Service of Instituto Fernandes Figueira and Hospital Universitário Gaffreé-Guinle, Rio de Janeiro, Brazil. Samples were stored in a freezer at -20°C. The Institutional Research Ethics Committee approved the study with consent of the children´s parents in all centers. We chose Down syndrome samples because there is an appropriate genetic marker and because there is indirect evidence that the leukemogenic process starts in utero (Gale et al., 1997). DNA extraction All DNA extraction and molecular analyses were performed at the Instituto Nacional de Câncer, Rio de Janeiro, Brazil. Cell suspensions were initially centrifuged to remove methanol and glacial acetic acid. These suspensions were then precipitated by centrifugation at 6000 rpm for 5 min, at room temperature. Supernatant was removed and cells were resuspended in 300 µL cold (4°C) PBS solution (NaCl, KCl, KH2PO4, NaHPO4, 12H2O, pH 7.2-7.4). This washing can be repeated twice or three times to remove all methanol/glacial acetic acid residues. Cell suspensions were lysed with 1.0 mL DNAzol reagent® (Invitrogen) by gently pipetting (Chomczynski et al., 1997). A 0.5-mL aliquot of 100% ethanol was added to the DNAzol cell lysate. This mixture was shaken vigorously and stored for 5 min at -80°. Then, centrifugations at 10,000 rpm for 5 min were performed to precipitate DNA. After centrifugation, the supernatant was removed and 1 mL 75% ethanol was added. The tube was agitated vigorously until the DNA pellet was completely dispersed. The resulting mixture was centrifuged at 12,000 rpm for 5 min at room temperature. Ethanol wash was decanted carefully and the tubes were stored vertically for approximately 1 min. Residual ethanol was removed with a micropipette. The DNA pellet was dissolved in 50 µL 8 mM NaOH and incubated at room temperature for 3-5 min, followed by repetitive pipetting. DNA was quantified with a NanoDrop®(ND-1000) spectrophotometer, with 1 µL of sample at 260 nm wavelength for determination of double-stranded DNA within a large dynamic range (2-3700 ng/µL DNA); the quality of the extraction was evaluated on an electrophoresis gel stained with ethidium bromide. To further test the quality of the DNA, a PCR for suspension cell DNA amplification was carried out with all samples in a 20-µL reaction volume. Each reaction tube contained at least 50 ng DNA; exon 2 of the GATA1 gene was amplified using primers described by Magalhães et al. (2006). After amplification, 5 µL PCR products was submitted to 1.5% agarose gel electrophoresis using a 100-bp ladder as a molecular weight marker. Denaturing high-performance liquid chromatography was performed on a Transgenomic WAVE System to screen for exon 2 of the GATA1 gene. RESULTS We were able to isolate DNA from samples that have been collected and stored for over three years. The amount of DNA varied from 36 to 305 ng/µL, with a mean of 100.2 ng/µL (N = 30). The A260/A280 values ranged from 1.5 to 2.7. The mean value of the A260/A280 ratio was 2.1, based on Sambrook et al. (1993). The amount and quality of DNA was suitable to amplify exon 2 of the GATA1 gene on a 1.5% agarose gel (Figure 1).
Sambrook J, Fritsch EF and Maniatis T (1993). Molecular cloning - a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. | |