Candida albicans GRX2, encoding a putative
glutaredoxin, is required for virulence in a
murine model
G.M. Chaves, S. Bates, D.M. MacCallum and F.C. Odds
Aberdeen Fungal Group, School of Medical
Sciences,
Institute of Medical Sciences, Aberdeen, UK
The present address of G.M. Chaves is Disciplina de Infectologia,
Departamento de Medicina, Universidade Federal de São Paulo, São Paulo,
SP, Brasil
Corresponding author: F.C. Odds
E-mail: [email protected]
Genet. Mol. Res. 6 (4): 1051-1063 (2007)
Received May 10, 2007
Accepted October 11, 2007
Published November 27, 2007
ABSTRACT. Resistance of Candida albicans to reactive oxygen species is thought to enhance its virulence in mammalian hosts. Genes such as SOD1, which encodes the anti-oxidant, superoxide dismutase, are known virulence factors. We disrupted the gene GRX2, which encodes a putative glutathione reductase (glutaredoxin) in C. albicans, and we compared the mutant with an sod1Δmutant. In vitro, the grx2Δstrain, but not the sod1Δstrain, was defective in hypha formation. The grx2Δstrain, but not sod1Δ, was significantly more susceptible to killing by neutrophils. When exposed to two compounds that generate reactive oxygen species, both mutants were susceptible to 1 mM menadione, but grx2Δnull alone was resistant to diamide. Both mutants were attenuated in a murine intravenous challenge model, and a GRX2 reintegrant regained partial virulence. Emphasis on the putative function of products of genes such as SOD1 and GRX2 in resistance to oxidative stress may oversimplify their functions in the virulence process, since the grx2Δstrain also gave defective hypha formation. Both mutants were sensitive to menadione and were slow to form germ tubes, though growth rates matched controls once the lag phase was passed.
Key words: Candida albicans, Virulence, GRX2, Glutathione reductase
INTRODUCTION
Candida albicans is the most common cause of life-threatening, opportunistic fungal disease in severely debilitated or immunocompromised patients (Calderone, 2002). It is a diploid fungus that can assume different morphologies, from budding yeast cells through pseudohyphae to true hyphae, depending on environmental conditions (Berman and Sudbery, 2002). More than 70 genes have now been implicated as contributors to the virulence of C. albicans in experiments based on selective gene disruption and on virulence testing by intravenous challenge in experimental mice (Navarro-Garcia et al., 2001; Brand et al., 2004).
The anti-Candida role of the oxidative burst within phagocytic leukocytes has long been recognized, along with observations indicating that this fungus possesses mechanisms to resist reactive-oxygen species (Odds, 1988). The C. albicans genome includes a family of six superoxide dismutase (SOD) genes (Martchenko et al., 2004) with the potential to convert damaging superoxide radicals to hydrogen peroxide, which can be detoxified by catalase. The cytosolic CuII/ZnII superoxide dismutases encoded by SOD1 (Hwang et al., 2002) and SOD5 (Martchenko et al., 2004) have been implicated as virulence factors in specific gene disruption studies. SOD5 expression is increased in C. albicans yeast cells exposed to polymorphonuclear neutrophils (PMN) (Fradin et al., 2005) and during morphological change from yeasts to hyphae (Nantel et al., 2002). However, while sod1Δis hypersensitive to killing by a macrophage cell line in vitro (Hwang et al., 2002), the same was not found for sod5Δ(Martchenko et al., 2004), indicating that the virulence role of Sod5p may be due to resistance to PMN but not to macrophages. MnII-dependent mitochondrial Sod2p in C. albicans has been ruled out as a putative virulence factor (Hwang et al., 2003), and the virulence roles of SOD3, SOD4 and SOD6 have not yet been directly investigated.
Exposure of C. albicans cells to low levels of hydrogen peroxide leads to an increase in catalase, glutathione reductase and superoxide dismutase activity, while higher levels of H2O2 are associated only with an increase in glutathione reductase activity (Gonzalez-Parraga et al., 2003). Transcript profiling of H2O2-stressed C. albicans showed an increase in expression of GRX1, GRX2 and CTA1, but not of SOD2 (Enjalbert et al., 2003), matching the enzyme data. Disruption of the catalase-encoding gene CTA1 generates a virulence-attenuated mutant (Wysong et al., 1988), and glutathione reductase activities have been repeatedly implicated as correlated not only with oxidative stress resistance but also with yeast-hypha morphogenesis (Gunasekaran et al., 1995), a property itself often regarded as a contributor to C. albicans virulence (Gow et al., 2002). However, a role for glutathione metabolism in C. albicans virulence has not been directly investigated. Glutaredoxins (glutathione-reductase enzymes) are ubiquitous cytosolic proteins that catalyze reduction of glutathione disulphide. Mammalian glutaredoxins are involved in a wide variety of cellular processes, including resistance to oxidative stress and growth control (Arnér and Holmgren, 2000); they may therefore play an adjunct role in defense against reactive oxygen species (ROS). They have been shown to play a role in combating oxidative stress in the erythrocytic stages of Plasmodium falciparum (Muller, 2004).
Like Saccharomyces cerevisiae, C. albicans possesses more than one gene homologue encoding glutaredoxin activity. One of these, presently named TTR1, is a homologue of GRX2 in S. cerevisiae. Expression profiling shows that this gene is upregulated not only in C. albicans cells exposed to H2O2 (Enjalbert et al., 2003; Gonzalez-Parraga et al., 2003), but also in cells exposed to the inhibitor benomyl (Karababa et al., 2004). Other glutaredoxin homologues are not upregulated in response to such stresses. When Fradin et al. (2005) compared C. albicans gene expression in response to PMN with whole blood depleted of PMN as a control, they found that expression of GRX2 was more highly upregulated than that of SOD1, under both circumstances. We, therefore, decided to investigate this particular gene as a putative virulence factor, possibly involved in defense against oxidative stress. The gene name TTR1 derives from the older yeast literature, from which the abbreviation related to thiol transferase and the name TTR1 carried over into the C. albicans literature and genome databases (http://www.candidagenome.org/, http://genolist.pasteur.fr/CandidaDB/). However, available genomic information indicates at least three homologues of glutathione reductase in C. albicans bearing the characteristic CP*C motif, two of which (orf19.6059 and orf19.3920) are homologous to S. cerevisiae GRX2. We, therefore, chose to name the gene formerly described as TTR1 (orf19.6059) as CaGRX2, since it is likely in the long term that GRX-based nomenclature will be used for the entire family of glutaredoxin-encoding genes. GRX2 is an acknowledged synonym of TTR1 (http://genolist.pasteur.fr/CandidaDB/).
We investigated the role of GRX2 in C. albicans by specific gene disruption and characterization of the mutant. We found that the grx2Δnull mutant is attenuated in mouse virulence and is more readily killed by PMN in vitro. An sod1Δmutant was susceptible to both menadione and diamide, which generate ROS in vitro; however, the grx2Δnull was menadione-susceptible and diamide-resistant, suggesting complexity in the cellular roles of individual anti-oxidant proteins.
MATERIAL AND METHODS
Strains, media and culture conditions
All C. albicans strains used and constructed in this study are listed in Table 1. They were stored at -80°C in 20% (v:v) glycerol and later subcultured on Sabouraud agar. The following media were used routinely to grow the fungi. NGY broth: 1 g/L Neopeptone (Difco, Detroit, MI, USA), 4 g/L glucose and 1 g/L yeast extract; YPD broth: 10 g/L yeast extract, 20 g/L glucose and 20 g/L Mycological Peptone (Oxoid, Basingstoke, UK); SD medium: 6.7 g/L Yeast Nitrogen Base with ammonium sulphate and without amino acids (Difco), 20 g/L glucose, with 25 μg/mL uridine added as required.

Construction of the grx2Δnull mutant and reintegrant strain
All primers used in this study are listed in Table 2. The GRX2 gene was disrupted by the ura-blaster method (Fonzi and Irwin, 1993). The regions of homology were amplified by polymerase chain reaction (PCR) using the upstream primer pair GRX21 fwd and GRX22 rev, containing HindIII and SphI restriction sites, respectively, and the downstream primer pair GRX23 fwd and GRX24 rev, containing Asp718 and BanII restriction sites, respectively. The 460-bp upstream and 370-bp downstream products were cloned into the complementary restriction sites in pMB-7 (Fonzi and Irwin, 1993). The disruption cassette was released by digestion with HindIII and BanII, and GRX2 was disrupted by sequential rounds of transformation into strain CAI-4. The URA3 marker was recycled by selection on SD medium plus 5-fluoroorotic acid (1 mg/mL) and uridine (50 μg/mL). Gene disruption was confirmed by PCR. To avoid potential problems associated with ectopic expression of URA3 (Brand et al., 2004), the Ura – grx2Δmutant was transformed with StuI-digested CIp10 plasmid (Murad et al., 2000), ensuring URA3 expression at the neutral RPS1 locus (orf19.3002). The Ura – sod1Δand sod2Δmutants (kindly provided by Dr. Sa-Ouk Kang from the Laboratory of Biophysics, Seoul National University, Republic of Korea) were also transformed with StuI-digested CIp10.

As a control, a reintegrant strain was constructed, in which the GRX2 gene was introduced into the grx2Δmutant. The GRX2 open-reading frame plus 934 bp of its promoter and 546 bp of its terminator was amplified by PCR (primers GRX2SF and GRX24 rev), and the product cloned into pGEM-T Easy (Promega Ltd., Southampton, UK). The plasmid insert was subcloned into the NotI site of CIp10. The resulting plasmid was digested with StuI and transformed into the Ura – grx2Δnull mutant.
RT-PCR for GRX2 expression
RNA was extracted by the method of Hayes et al. (2002), and multiplex semi-quantitative RT-PCR was done as previously described (Copping et al., 2005). Primers GRX2 fwd and GRX2 rev and EFB1 fwd and EFB1 rev were used to amplify GRX2 and EFB1 cDNA, respectively. EFB1 contains an intron and was used as both a control for genomic DNA contamination and as an expression standard. PCR products were sampled after every two amplification cycles, from cycle 12 to cycle 28, and GRX2 gene expression was estimated relative to EFB1.
Candida albicans growth rate determination
Growth rates were measured in duplicate. A volume from an overnight culture in NGY was transferred to YPD broth or RPMI 1640 (Gibco, Paisley, UK) to give an initial OD600 nm = 0.05 and the culture incubated at 37°C rotated at 200 rpm in a gyratory shaker. Growth was measured by OD600 nm versus a medium blank. Maximum growth rates were determined from the logarithms of values taken in the exponential phase.
Tests for hypha formation
For induction of hypha formation on solid media, the cells were grown in NGY, centrifuged and washed three times in water. From a suspension adjusted to 109 cells/mL, 5 μl was spotted on the surface of Spider (Liu et al., 1994) and GlcNAc agars (20 g/L N-acetyl-D-glucosamine, 6.7 g/L Yeast Nitrogen Base, 16 g/L Micro agar) in triplicate. The plates were incubated for seven days at 30°C.
For tests in liquid media, cells were grown, washed and standardized to an initial concentration of 106 cells/mL in 10% fetal calf serum (FCS) or YPD + 10% FCS pre-warmed to 37°C and incubated at the same temperature, with gyratory shaking at 200 rpm. After 1- and 3-h incubation, samples of the cultures were mixed with an equal volume of 10% formaldehyde to arrest further development. The sample at 1 h was examined microscopically for determination of the percentage of cells bearing evaginations. The 3-h sample was examined for approximation of the mean morphology index (MI) (Merson-Davies and Odds, 1989), in which a value close to 1 indicates a population of spheroidal yeast cells and a value close to 4 indicates a population of true hyphal cells, with values between 1 and 4 indicating mixed or pseudohyphal morphologies (Odds et al., 2000).
Proteinase assay
Proteinase activity was determined by a method modified from Macdonald and Odds (1980). Fifty-microliter samples from NGY cultures were grown in 5 mL YCB + BSA medium (11.7 g/L Yeast Carbon Base [Difco]; 10 g/L glucose; 5 g/L bovine serum albumin, fraction V, Batch 08k0560 [Sigma]) rotated in a wheel angled 5° from the horizontal at 30°C for 72 h. Proteolytic activity was determined by measuring the increase in trichloroacetic acid soluble products absorbing at 280 nm in triplicate after 1-h incubation of the culture supernatant with BSA substrate at 37°C. Specific activity was expressed as OD280/OD600 of the culture.
Phospholipase assay
Phospholipase activity was estimated by the egg yolk agar method (Price et al., 1982), with inocula prepared from overnight NGY cultures standardized to 2 x 105 cells/mL.
Candida albicans adhesion assay
The method of Kimura and Pearsall (1978) was used; C. albicans cells were grown overnight to stationary phase in SD at 37°C and were mixed with human buccal epithelial cells (HBEC) from healthy volunteers at a ratio of 1000 yeast cells per HBEC. The mixtures were incubated at 37°C for 45 min with shaking; then cells were filtered on polycarbonate (12 µm pores), Gram-stained and washed before transfer to a microscope slide. The number of C. albicans cells adhering to 100 HBEC was determined with the operator blinded to the nature of the material on the slide. Tests were done in triplicate.
Sensitivity of Candida albicans to oxidative-stress inducers
The method of Izawa et al. (1995) was used with some modifications to determine the sensitivity of C. albicans to oxidative-stress inducers. Yeasts grown in NGY were standardized to 2 × 107 cells/mL. Five-microliter volumes of a 10-fold dilution series prepared from this suspension were spotted on the surface of YPD agar plates containing concentrations of menadione and diamide ranging from 0.5 to 2.5 mM. The plates were incubated for 48 h at 30°C. Alternatively, 50-μl volumes of NGY-grown yeasts were transferred to 5 ml YPD broth, with and without additions of menadione or diamide (0.5-2.0 mM), incubated in a rotator wheel for 16 h at 30°C and the OD600 nm determined for control and test tubes. Sensitivity to the compounds was measured by examining growth in the presence of compounds as a percentage of control growth. Sensitivity to H2O2 was tested in the same way, except that incubation was continued for 3 h instead of 16 h.
Candida albicans killing by polymorphonuclear neutrophils
PMN freshly isolated from blood samples of healthy volunteers on the day of the experiment (Fradin et al., 2005) were suspended in Eagle’s minimal essential medium (Gibco) + 20 mM HEPES, pH 7.2, and standardized to 8 x 105 PMN/mL. C. albicans cells grown overnight in NGY were centrifuged and washed three times in saline and resuspended at 5 × 106 yeasts/mL in HEPES-buffered Eagle’s minimal essential medium containing one-tenth volume of fresh human plasma. Equal volumes of PMN and yeast suspensions were mixed and incubated at 37°C for 1 h with rotation at 50 rpm. Control suspensions contained C. albicans without PMN. The suspensions were centrifuged and the pellets resuspended in water to lyse the PMN. After three cycles of washing and resuspension, viable C. albicans counts were determined by plating a 10-fold dilution series on YPD agar. Killing was determined as the mean difference between viable counts in the presence and absence of PMN. The assays were performed in triplicate.
Virulence of Candida albicans in the murine IV challenge model
Animal experimentation was done under the terms of UK Home Office licenses for research on animals. Immunocompetent female BALB/c mice (Harlan Sera-lab, Loughborough, UK) with a weight range from 17-23 g were supplied with food and water ad libitum. Mice were intravenously inoculated with C. albicans strains that had been grown overnight in NGY, washed and resuspended in saline. The inoculum was standardized to allow injection of 2-3 × 104 cfu/g mouse body weight. Mice were examined daily and animals showing a loss of body weight >20% or which showed signs of serious illness were humanely terminated and kidneys and brain removed aseptically for determination of fungal burdens by viable counting.
Statistical analysis
Means ± standard deviations were determined from the results of at least three independent experiments. Differences between values for phenotypic tests were analyzed by the Student t-test. Animal survival data were compared with Kaplan-Meier/LogRank statistics, and differences between tissue burden data with the Mann-Whitney U-test. P values <0.05 were considered to be significant for all comparisons.
RESULTS
Gene disruptions and reintegrations
GRX2/TTR1 was identified in the CandidaDB C. albicans genomic database as orf19.6059. BLAST searching revealed orf19.3920 as a close homologue, also annotated as encoding a putative glutaredoxin and even listed as a synonym of GRX2/TTR1. The nomenclature of the GRX gene family in C. albicans is presently a source of confusion, but our study was based on the sequence of orf19.6059, and primers for GRX2 were designed according to this DNA sequence.
Table 1 details the mutants generated in this study. The grx2Δmutant GCY206 was made by deletion of 1583 bp from the central region of the GRX2 gene in C. albicans CAI-4 with the “Ura-blasting” technique (Fonzi and Irwin, 1993). The URA3 gene was reintegrated at the neutral RPS1 locus, to avoid ectopic URA3 expression problems (Brand et al., 2004), creating mutant GCY207. The GRX2 gene as well as URA3 were integrated at the RPS1 locus of the grx2Δnull mutant to make the grx2Δ+ GRX2 reintegrant GCY208. The URA3 gene was also reintegrated at the RPS1 locus in sod1Δ and sod2Δ mutants to create mutants GCY200 and GCY201, respectively.
Phenotypic properties of the mutants in vitro
Growth rates of yeast cells of all the null mutants and the GRX2 reintegrant strain were essentially the same as for the wild-type NGY152 (Table 3). Putative virulence properties, such as proteinase, phospholipase activity and adhesion to HBEC were unaltered in the mutants and reintegrant strains. Colony morphologies on Spider and GlcNAc agars were not markedly different for the mutants when compared to NGY152.
Around 90% of NGY152 yeast cells had evaginated within 1 h in 10% FCS and YPD + FCS (Table 3). The sod1Δ and sod2Δ null mutants evaginated more slowly in YPD + FCS, but did so at the same rate in 10% serum. The grx2Δ mutant, GCY207, and the reintegrant, GCY208, were markedly delayed in evagination at 1 h, particularly in YPD + FCS.
After 3 h of incubation in 10% FCS, NGY152 cells formed true hyphae (mean MI = 4; Table 3) and the mean MI for the mutants similarly suggested formation of true hyphae in serum (Table 3). In YPD + FCS, while NGY152 cells continued to show a hyphal morphology (mean MI of 3.9), all mutants and the GRX2 reintegrant had a lower mean MI, often with a large standard deviation (Table 3), suggestive of mixed morphologies with reduced proportions of true hyphae. The grx2Δ mutant, GCY207, was particularly deficient in hypha formation in YPD + FCS, with a mean MI of only 1.3, indicating growth mainly in the yeast form or as short pseudohyphae.
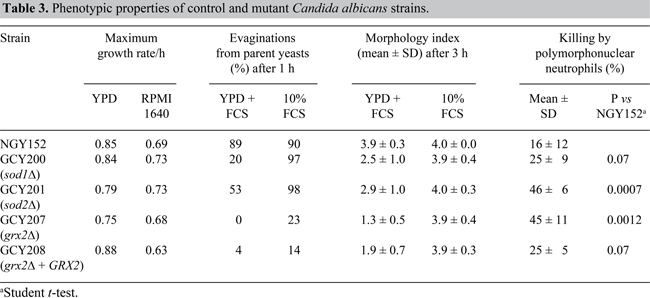
Effects of oxidative stress inducers
All mutants and the GRX2 reintegrant were more susceptible than NGY152 to menadione (Figure 1A). GCY201 (sod2Δ) was the most susceptible to menadione. Agar plate tests gave essentially similar results (data not shown). A different pattern of susceptibility was observed with diamide (Figure 1B). In the presence of 2 mM diamide, GCY207 (grx2Δ) grew almost to control levels, while every other strain was inhibited. In plate tests, growth of GCY207 was unaffected even at 2.5 mM diamide, whereas NGY152 was completely inhibited at this concentration. All the strains were equally sensitive to H2O2, with growth reduced below 50% of control by 2.5 mM H2O2 after 3-h incubation (data not shown).
Effects of polymorphonuclear neutrophils
The grx2Δ and sod2Δ null mutants were significantly more susceptible to killing by PMN than NGY152 (Table 3) but the increase in killing measured for the sod1Δ null (GCY200) and the GRX2 reintegrant (GCY208) was not significantly different from control values.
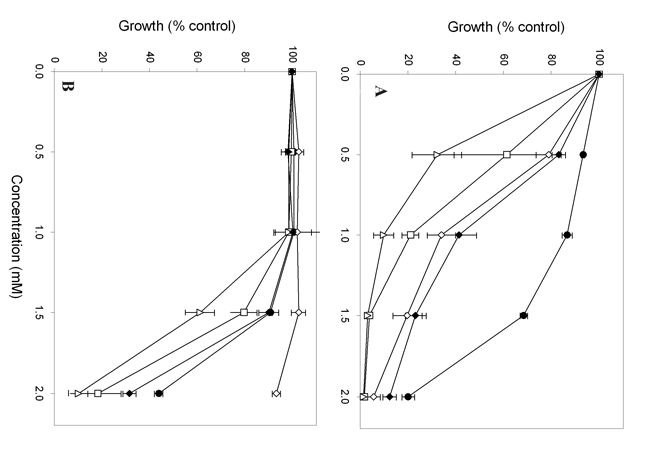
Virulence of mutants in the murine IV challenge model
Median survival times for animals infected with GCY200 (sod1Δ) or GCY207 (grx2Δ) was 28 days, as compared with five days for the control strain NGY152 (LogRank test; p < 0.001; Figure 2). The survival curve for the GRX2 reintegrant strain GCY208 indicated some regain of virulence, but not to control levels (p < 0.001 relative to NGY152). Viable count data showed significantly reduced tissue burdens in both kidneys and brain for mice infected with null mutants GCY200 and GCY207. Kidney burdens were not significantly different from control for mice infected with the GRX2 reintegrant GCY208, but brain burdens were significantly lower (Table 4).
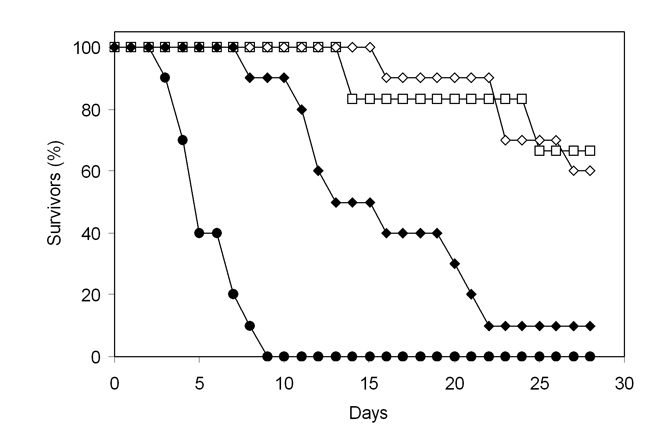
Figure 2. Survival of mice challenged intravenously with Candida albicans strains. Closed circles, NGY152; open squares, GCY200 (sod1Δ); open diamonds, GCY207 (grx2Δ); closed diamonds, GCY208 (grx2Δ+ GRX2).
DISCUSSION
A possible role for C. albicans GRX2 in virulence has been directly demonstrated in this study by the observations that survival was extended (Figure 2) and mean tissue burdens reduced (Table 4) in animals challenged with a grx2Δ mutant. However, reintegration of a single copy of GRX2 in the null mutant partly, but not wholly, reversed virulence attenuation, and did not fully restore wild-type phenotypic properties in vitro. Similar inexplicable failures of reintegrants to regain full wild-type virulence have been noted for other genes, including some regarded as encoding virulence factors in similarly designed experiments (Hobson et al., 2004; Palmer et al., 2004; Tripathi et al., 2004; Bates et al., 2005) presumably because reintegration of only one copy of these genes is not enough for fully regaining of wild-type phenotype. We measured the level of expression of GRX2 by semi-quantitative multiplex RT-PCR and found no difference between levels in the reintegrant, GCY208, and control, NGY152 (details not shown); therefore, expression level differences cannot account for the failure of the reintegrant to recover wild-type properties. The survival and burden data for the grx2Δ mutant were very similar to results for the sod1Δ mutant. We tested sod1Δ in this study with URA3 reintegrated at the highly expressed RPS1 locus to avoid artefacts of URA3 underexpression (Brand et al., 2004); the results confirm the original report of virulence attenuation in this mutant (Hwang et al., 2002).
Although the grx2Δ and sod1Δnulls were attenuated in mouse virulence, their roles in the pathogenesis process cannot be similar. The sod1Δmutant was unaffected in hypha formation in YPD + FCS and in 10% FCS alone, while the grx2Δmutant had a delay in the evagination of daughter cell material under both conditions; there was a preponderance of yeast cells and short pseudohyphae in YPD + FCS (Table 3). Killing of the grx2Δ null by PMN was significantly increased over that of the control, while killing of the sod1Δnull was not. The sod1Δnull was inhibited by 1 mM menadione and 2 mM diamide, while the grx2Δnull was resistant to 2 mM diamide, indicating differences in the roles of SOD1 and GRX2 in combating oxidative stress.
Pereira et al. (2003) demonstrated that in S. cerevisiae the sod2Δmutant is not sensitive, based on survival of the cells after exposure to menadione. They suggested that SOD1 is sufficient to provide ROS tolerance in an sod2Δmutant. Based on our study, together with the findings of Hwang et al. (2002, 2003), we infer that the role of SOD genes is different in C. albicans and S. cerevisiae because the C. albicans sod2Δmutant is very sensitive to menadione, emphasizing a greater importance of this enzyme in ROS resistance to menadione-induced stress than in S. cerevisiae.
Susceptiblity of the sod1Δmutant to diamide in C. albicans has not been investigated previously. Diamide is a thiol-specific oxidant that can readily oxidize glutathione (O’Brien, 1970; Kosower and Kosower, 1995). Therefore, a difference in diamide susceptibility between the sod1Δmutant and the wild type is not expected.
We investigated the role of glutaredoxin in C. albicans. Apparently, menadione plays a minimal role in oxidative stress in the grx2Δ mutant. It is not surprising that the stress generated by menadione does not make grx2Δ very susceptible, because glutaredoxins do not interact directly with O2-.
The C. albicans grx2Δ mutant was very resistant to diamide. If this chemical is a rapid thiol oxidant, then this mutant should be sensitive rather than resistant to diamide. The resistance of the Δgrx2 mutant that we found could be due to a compensatory mechanism of the other glutaredoxins present in C. abicans. The situation is clearly complicated, and extensive specific study may be required to evaluate the individual roles of all S-thiolation and oxidative-stress responses in C. albicans.
It is possible that the virulence attenuation found in the grx2Δ null was a consequence of changes in properties other than resistance to oxidative stress. With families of both SOD and GRX genes in C. albicans it is possible that redundancy allows other members of the family to take over the putative oxidative-stress resistance roles of SOD1 or GRX2, when either is disrupted singly. Phenotypic behavior of the sod2Δ mutant closely followed that of the sod1Δ null, yet this mutant is not attenuated in virulence for mice (Hwang et al., 2003).
Attenuation in mouse virulence in the grx2Δ null might be related to its slower evagination in two growth media, and its defective hypha formation in YPD + FCS, as well as its susceptibility to neutrophil killing (Table 3). However, the sod2Δ null was also significantly more vulnerable than NGY152 to PMN killing, yet it is reportedly normally virulent for mice (Hwang et al., 2003). We did not directly retest the sod2Δ null for mouse virulence, since it is unethical to further expose animals to challenge with a strain already shown to possess wild-type virulence.
We have extended the rapidly growing list of genes whose disruption leads to virulence attenuation in the mouse challenge model. The differences in effects of diamide on grx2Δ and sod1Δ mutants suggest complex patterns of regulation of different C. albicans genes; emphasis of their properties in resistance to oxidative stress may oversimplify their virulence functions in vivo.
ACKNOWLEDGMENTS
We thank the Brazilian Ministry of Education (CAPES) for financial support to G.M. Chaves. S. Bates and D.M. MacCallum were supported by grants from the Wellcome Trust. We are grateful to Dr. Sa-Ouk Kang for the sod1Δ and sod2Δ strains.
REFERENCES
Arnér ES and Holmgren A (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267: 6102-6109.
Bates S, MacCallum DM, Bertram G, Munro CA, et al. (2005). Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+- ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280: 23408-23415.
Berman J and Sudbery PE (2002). Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3: 918-930.
Brand A, MacCallum DM, Brown AJ, Gow NA, et al. (2004). Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3: 900-909.
Calderone RA (2002). Candida and candidiasis. ASM Press, Washington.
Copping VM, Barelle CJ, Hube B, Gow NA, et al. (2005). Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55: 645-654.
Enjalbert B, Nantel A and Whiteway M (2003). Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell. 14: 1460-1467.
Fonzi WA and Irwin MY (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134: 717-728.
Fradin C, De Groot P, MacCallum D, Schaller M, et al. (2005). Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56: 397-415.
Gonzalez-Parraga P, Hernandez JÁ and Arguelles JC (2003). Role of antioxidant enzymatic defences against oxidative stress H(2)O(2) and the acquisition of oxidative tolerance in Candida albicans. Yeast 20: 1161-1169.
Gow NA, Brown AJ and Odds FC (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5: 366-371.
Gunasekaran S, Imbayagwo M, McDonald L, Gunasekaran M, et al. (1995). Influence of carbon and nitrogen sources on glutathione catabolic enzymes in Candida albicans during dimorphism. Mycopathologia 131: 93-97.
Hayes A, Zhang N, Wu J, Butler PR, et al. (2002). Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26: 281-290.
Hobson RP, Munro CA, Bates S, MacCallum DM, et al. (2004). Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279: 39628-39635.
Hwang CS, Rhie GE, Oh JH, Huh WK, et al. (2002). Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148: 3705-3713.
Hwang CS, Baek YU, Yim HS and Kang SO (2003). Protective roles of mitochondrial manganese-containing superoxide dismutase against various stresses in Candida albicans. Yeast 20: 929-941.
Izawa S, Inoue Y and Kimura A (1995). Oxidative stress response in yeast: effect of glutathione on adaptation to hydrogen peroxide stress in Saccharomyces cerevisiae. FEBS Lett. 368: 73-76.
Karababa M, Coste AT, Rognon B, Bille J, et al. (2004). Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48: 3064-3079.
Kimura LH and Pearsall NN (1978). Adherence of Candida albicans to human buccal epithelial cells. Infect. Immun. 21: 64-68.
Kosower NS and Kosower EM (1995). Diamide: an oxidant probe for thiols. Methods Enzymol. 251: 123-133.
Liu H, Kohler J and Fink GR (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723-1726.
MacDonald F and Odds FC (1980). Inducible proteinase of Candida albicans in diagnostic serology and in the pathogenesis of systemic candidosis. J. Med. Microbiol. 13: 423-435.
Martchenko M, Alarco AM, Harcus D and Whiteway M (2004). Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell. 15: 456-467.
Merson-Davies LA and Odds FC (1989). A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 135: 3143-3152.
Muller S (2004). Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 53: 1291-1305.
Murad AM, Lee PR, Broadbent ID, Barelle CJ, et al. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16: 325-327.
Nantel A, Dignard D, Bachewich C, Harcus D, et al. (2002). Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 13: 3452-3465.
Navarro-Garcia F, Sanchez M, Nombela C and Pla J (2001). Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25: 245-268.
O’Brien RW, Weitzman PD and Morris JG (1970). Oxidation of a variety pf natural electron donors by the thiol-oxidising agent, diamide. FEBS Lett. 10: 343-345.
Odds FC (1988). Candida and candidosis. 2nd edn. Bailliere Tindall, London.
Odds FC, Van Nuffel L and Gow NA (2000). Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146: 1881-1889.
Palmer GE, Johnson KJ, Ghosh S and Sturtevant J (2004). Mutant alleles of the essential 14-3-3 gene in Candida albicans distinguish between growth and filamentation. Microbiology 150: 1911-1924.
Pereira MD, Herdeiro RS, Fernandes PN, Eleutherio EC, et al. (2003). Targets of oxidative stress in yeast sod mutants. Biochim. Biophys. Acta 1620: 245-251.
Price MF, Wilkinson ID and Gentry LO (1982). Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 20: 7-14.
Tripathi G, Wiltshire C, Macaskill S, Tournu H, et al. (2004). Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21: 5448-5456.
Wysong DR, Christin L, Sugar AM, Robbins PW, et al. (1988). Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66: 1953-1961.
